Abstract
Expansion of an unstable GAA·TTC repeat in the first intron of the FXN gene causes Friedreich ataxia by reducing frataxin expression. Deficiency of frataxin, an essential mitochondrial protein, leads to progressive neurodegeneration and cardiomyopathy. The degree of frataxin reduction correlates with GAA·TTC tract length, but the mechanism of reduction remains controversial. Here we show that transcription causes extensive RNA·DNA hybrid formation on GAA·TTC templates in bacteria as well as in defined transcription reactions using T7 RNA polymerase in vitro. RNA·DNA hybrids can also form to a lesser extent on smaller, so-called ‘pre-mutation’ size GAA·TTC repeats, that do not cause disease, but are prone to expansion. During in vitro transcription of longer repeats, T7 RNA polymerase arrests in the promoter distal end of the GAA·TTC tract and an extensive RNA·DNA hybrid is tightly linked to this arrest. RNA·DNA hybrid formation appears to be an intrinsic property of transcription through long GAA·TTC tracts. RNA·DNA hybrids have a potential role in GAA·TTC tract instability and in the mechanism underlying reduced frataxin mRNA levels in Friedreich Ataxia.
INTRODUCTION
RNA·DNA hybrid formation is most often associated with DNA replication, where a role for the hybrid as the primer for DNA synthesis is well known (1). Other uses are less clear, and cells possess a host of activities to prevent or remove the hybrids, suggesting that RNA·DNA hybrids may not be generally beneficial. For instance, even simple RNA polymerases such as T7 RNAP separate the template from the nascent RNA, which threads out of the polymerase through a positively charged exit pore (2,3). In addition, multi-subunit RNA polymerases such as that of Escherichia coli have a ‘lid’ element that functions to separate RNA from the RNA·DNA hybrid in the active site, guiding it to the exit pore and inhibiting the formation of long RNA·DNA hybrids even on single-stranded templates (4,5).
RNase H activities that specifically digest the RNA in an RNA·DNA hybrid are ubiquitous in both prokaryotes and eukaryotes (6). Also widely expressed are topoisomerases, which can remove the negative supercoils that favor RNA·DNA hybrid formation (7). Topoisomerase activity may be particularly crucial to reducing RNA·DNA hybrid formation because a domain of negative supercoiling is generated behind a transcribing RNA polymerase (8,9). Ribosomes coat the nascent transcript during co-transcriptional translation in bacteria and a faster RNA polymerase such as T7 will outrun the ribosomes leaving the transcript open to RNase digestion (10). Bound ribosomes may also discourage re-annealing to the template as translation inhibitors can increase RNA·DNA hybrid formation in bacteria (11). In eukaryotes, several different co-transcriptional systems may inhibit re-annealing by coating the nascent transcript as part of their functions in mRNP maturation. Furthermore, chromatin would further limit the accessibility of template DNA to RNA after transcription is completed.
In yeast, the THO/TREX system couples transcription elongation to mRNA export (12). The THO complex associates with sites of active transcription and recruits mRNA export proteins such as Sub2 and Yra1 to the nascent transcript (13,14). Many components of the THO/TREX complex are conserved between yeast and higher eukaryotes, but the order of loading is not. Unlike yeast, where the THO complex associated with the transcribing polymerase recruits splicing factors (14), the metazoan THO/TREX complex is itself recruited to mRNA during splicing by serine-arginine-rich (SR) splicing factors (15).
Breakdowns in co-transcriptional RNA processing due to mutations affecting THO/TREX can lead to RNA·DNA hybrid formation, impaired transcription elongation and increased genomic instability in yeast (16–18). Depletion of an SR protein called alternative splicing factor/splicing factor 2 (ASF/SF2) in the avian B-cell line DT40 will also cause RNA·DNA hybrid formation and genome instability (19). In both cases, RNA·DNA hybrids link impaired co-transcriptional RNA–protein interactions to genomic instability (18,19).
Some sequences have an intrinsic propensity to form RNA·DNA hybrids that may overcome co-transcriptional mechanisms repressing hybrid formation. For example, immunoglobulin class switch regions have long been known to form RNA·DNA hybrids when transcribed in vitro. Notably, the hybrids only form if the sequences are transcribed in the physiological direction to make a purine-enriched transcript (20,21).
The most common inherited ataxia, Friedreich ataxia (FRDA), is caused by expansion of an unstable GAA·TTC trinucleotide repeat within the first intron of the frataxin gene (FXN) (22). There is a direct relationship between the length of the expanded repeats and disease severity (23,24), reflecting reduced frataxin expression. However, the causes of GAA·TTC repeat instability are poorly understood and the means by which expanded repeats suppress gene expression are controversial (25–30). Most FRDA patients have intact frataxin-coding sequences, so a refined understanding of factors contributing to the transcript deficit and the repeat instability will aid in the design of therapies.
We have previously demonstrated that a long GAA·TTC tract forms a structural impediment to T7 RNA polymerase (28,31). Here we show that GAA·TTC tracts have an intrinsic length-dependent propensity towards RNA·DNA hybrid formation during transcription, both in vitro and in living bacteria cells. Furthermore, we show that an extensive RNA·DNA hybrid is tightly linked to T7 RNA polymerase arrest on templates that contain long GAA·TTC repeats.
MATERIALS AND METHODS
Plasmids
Uninterrupted (CAG·CTG)88 and (GAA·TTC)n repeats were made by defined stepwise expansion of smaller units cloned into a plasmid (pREX) designed for that purpose. Asymmetric type IIS restriction digestion, fragment purification and ligation to achieve that expansion has been described (32). Plasmids bearing a self-cleaving ribozyme 3′ to (CAG·CTG)88 and (GAA·TTC)88 repeats have been described (28). An SpeI–XbaI fragment was excised from pREX(GAA)88 and inserted into pBAD18 to make pBAD18(GAA)88. Plasmids were grown, and in vivo experiments were carried out in bacterial strain XL-1 Blue (Stratagene).
In vivo transcription
Vectors for bacterial transcription were under control of the arabinose inducible BAD promoter (33). Bacterial strain XL-1 Blue (Stratagene) containing plasmids with zero repeats (pBAD18) or 88 GAA·TTC repeats (pBAD18GAA88) were grown in parallel to mid-log phase. Glucose or arabinose was added to 0.2% for 20 min. Cells were collected by centrifugation at 37°C in pre-warmed holders. RNA and plasmid were rapidly isolated using the GTC-acid phenol method (34). Samples were treated with the single-strand specific RNases A and T1 or with the combination of RNases A&T1 and RNase H for 1 h at 32°C. Samples were resolved on a 1% agarose gel and stained with ethidium bromide after electrophoresis.
In vitro transcription
RNA transcription from phage promoters was performed in T7 transcription buffer (50 mM HEPES, pH 8.0, 100 mM NaCl, 20 mM MgCl2, 10 mM DTT and 0.5 mM each NTP) supplemented with 200 U/ml RNase Inhibitor (Ambion) in a final volume of 20 μl at 37°C for 20 min. T7 RNA polymerase (Ambion) was used at a final concentration of 1000 U/ml. Template concentrations (usually ∼200 ng/reaction) were estimated by ethidium bromide fluorescence. The 5′ end of the transcript was labeled by including [γ-32P]-GTP (6000 Ci/mmol). The gamma phosphate is retained only in the first position.
RNase digestion
RNase A and T1 (pre-mixed, Ambion) were used at a concentration of 20 μg/ml and 1000 U/ml, respectively, for 1 h at 37°C in TE (10 mM Tris HCl, pH 8.0, 1 mM EDTA) unless otherwise noted. RNase H (US Biochemical Corp.) digestion was performed in the buffer supplied by the manufacturer in the experiments where digestion followed transcription. In most cases, RNase H digestion was performed simultaneously with transcription in the T7 transcription buffer. To make an RNA marker ladder, RNase T1 (Ambion) was titrated on end-labeled transcripts containing (GAA)44 to obtain an appropriate partial digest.
Primer extension
An oligodeoxyribonucleotide with the sequence (5′-TGGACGAGTCTCGAGCAGCTGAAGCTTGCA-3′) that anneals from 71 to 40 bases 3′ of the end of the TRS in transcripts produced by T7 RNAP was synthesized using standard phosphoramidite chemistry (Life Technologies). The oligonucleotide (2 pmol) was end-labeled with [γ32P]GTP (6000 Ci/mmol, PerkinElmer) using T4 polynucleotide kinase (New England Biolabs) in the supplied buffer.
RNA was prepared for primer extension by performing transcription reactions with T7 RNAP on a supercoiled template containing (GAA·TTC)88 in the presence (2.0 U) or absence of RNase H for 1 h at 37°C in a volume of 25 μl. The reactions were stopped by adding 200 μl stop buffer (5 mM EDTA, 0.5% SDS, 0.2 mg/ml Proteinase K and 10 μg/ml tRNA) followed by incubation at 65°C for 20 min. The RNA was then extracted with 200 μl phenol:chloroform:isoamyl alcohol (25:24:1), followed by ethanol precipitation.
Primer extension was performed by taking aliquots of the purified RNA resuspended in H2O (10 μl) and adding 2 μl of dilute kinased oligonucleotide (∼0.1 pmol) followed by heating to 70°C for 10 min and then chilled on ice. The Superscript II, RNase H-reverse transcriptase first strand synthesis kit (Life Technologies) was then used following the manufacturers’ protocol.
Gel electrophoresis
Agarose gel electrophoresis was routinely performed in 1% agarose gels in TAE buffer (40 mM Tris acetate, 2 mM EDTA at pH 8.0). Gels were stained with ethidium bromide after electrophoresis. Radiolabeled reactions were stopped by mixing with 50 μl of stop buffer (98% deionized formamide, 10 mM EDTA and 10 μg/ml tRNA). The samples were ethanol precipitated, resuspended in a denaturing gel loading buffer (98% formamide, 10 mM EDTA and 0.05% bromophenol blue and xylene cyanole) heated to 90°C, and loaded on a pre-warmed 6% polyacrylamide sequencing gel containing 8 M urea. End-labeled pBR322 MspI digest (New England Biolabs) and 10 bp ladder (Life Technologies) were used as size markers. Images of radioactive gels were obtained using FujiFilm type BAS III-S phosphorimaging screens and a FujiFilm BAS 1500 reader. Analysis and quantitation was performed with FujiFilm Image Gauge 3.0 (Mac) software.
RESULTS
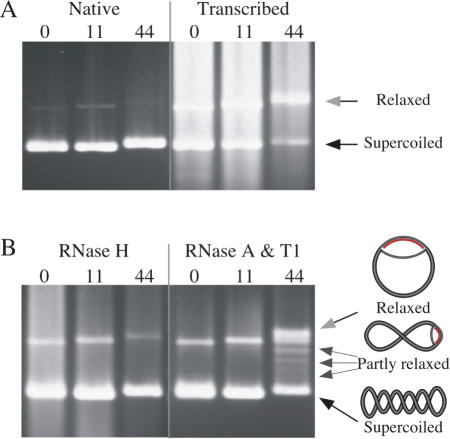
To investigate how expanded GAA·TTC tracts reduce gene expression we constructed a series of plasmids containing various lengths of the repeat and carried out transcription reactions on these templates in vitro. We noted that supercoiled plasmid templates with moderately long GAA·TTC tracts exhibit a more relaxed mobility in agarose gels after transcription if the product transcript contained a GAA tract. For instance, in Figure 1A, the native gel mobility of supercoiled plasmids carrying 0, 11 or 44 GAA·TTC repeats are shown in the first three lanes. Following transcription by phage T7 RNA polymerase (Figure 1A, last three lanes), the templates are partially obscured by transcripts, but it is clear that the template with 44 triplet repeats has shifted to a more relaxed mobility (gray arrowhead) rather than supercoiled mobility (black arrow).
Figure 1.
Transcription through a long GAA·TTC tract results in formation of an RNA·DNA hybrid. (A) The native gel mobility of supercoiled templates carrying 0, 11 or 44 GAA·TTC triplets is shown in the first three lanes and after transcription by T7 RNAP in the second three lanes. The RNA product partially obscures the templates. Gel mobilities of relaxed plasmids (gray arrowhead) and supercoiled plasmids (black arrowhead) are indicated. (B) Treatment with RNase H after transcription (first three lanes) returns the (GAA·TTC)44 template to control mobility. Treatment with the single-strand-specific RNases A and T1 (last three lanes) reveals conformers of the (GAA·TTC)44 template (small arrows) with mobilities approaching that of a fully relaxed template (gray arrowhead). The degree of relaxation reflects the length of the RNA·DNA hybrid, which unwinds negative supercoils as indicated in the schematic to the right of the arrows. In contrast, templates with 0 or 11 triplets retain the mobility of untranscribed controls, regardless of treatment.
Treatment of the transcribed samples with RNase H, which specifically degrades the RNA in an RNA·DNA hybrid, returns the transcribed (GAA·TTC)44 template to supercoiled mobility (Figure 1B, lane 3) indicating that the source of the mobility change is an RNA·DNA hybrid and not a nick in the template. Conversely, treatment with the single-strand-specific RNases A and T1 after transcription (Figure 1B, lanes 4–6) does not return the (GAA·TTC)44 template to supercoiled mobility, but reveals the appearance of a series of partially relaxed conformers (small arrows). Unwinding of the strand displaced by the R-loop relaxes negative supercoils, and the degree of relaxation reflects the length of the RNA·DNA hybrid (shown schematically to the right in Figure 1B). Different lengths of hybrid remain after single-strand-specific digestion partly because random branch migration can make single-stranded RNA ends available to the nucleases. The examples shown were treated with RNases A and T1 in low salt (TE), which provides a greater range of conformers than treatment in high salt which provides greater RNase protection (data not shown). The templates with 0 or 11 GAA·TTC repeats migrate with mobility expected of supercoiled templates after all treatments.
The samples were extracted with phenol prior to electrophoresis, so the altered mobility of the (GAA·TTC)44 template is unlikely to represent preferential binding of T7 RNA polymerase or RNase A or T1 to the repeat tract. Moreover, neither transcription of these same templates in the opposite direction by SP6 RNA polymerase, nor transcription of templates containing GAA·TTC repeats cloned in the opposite direction by T7 RNA polymerase produced RNA·DNA hybrids (data not shown). The appearance of relaxed conformers that are resistant to single-strand-specific RNases, but resolved by RNase H indicates the presence of an rGAA·dTTC hybrid.
A persistent rGAA·dTTC hybrid forms in bacteria
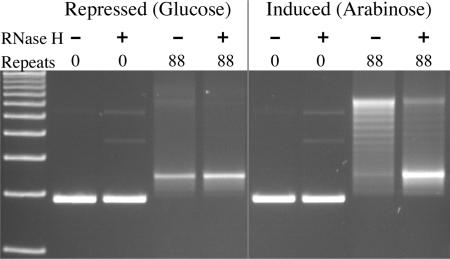
To verify that RNA·DNA hybrid formation in the GAA·TTC repeat was not restricted to in vitro transcription with simple phage RNA polymerases, we extended our analysis to transcription in bacteria. Plasmids were made with either 0 or 88 GAA·TTC repeats downstream of the sugar-regulated BAD promoter, which is induced by arabinose and repressed by glucose (33). Transcription of these plasmids was induced (or kept repressed) in live cells. The plasmids were then rapidly purified at 37°C to avoid the enhanced formation of RNA·DNA hybrids that may occur at lower temperatures (35). The plasmids were subsequently analyzed for RNA·DNA hybrids by gel electrophoresis, as in Figure 2.
Figure 2.
Persistent RNA·DNA hybrids form during transcription of GAA·TTC repeats in bacteria. The gel pictured shows the mobility of templates isolated from bacteria in which transcription was repressed by glucose (first 4 lanes) or induced by arabinose (last 4 lanes). The plasmids were treated after isolation with the single-strand-specific RNases A and T1. Some aliquots were additionally treated with RNase H as indicated. The transcribed templates with 88 GAA·TTC triplets show a fully or partially relaxed mobility when treated only with single-strand-specific RNases (next to last lane). Treating an aliquot of the same sample with RNase H returns the bulk of the plasmid to supercoiled mobility in the last lane.
The results shown in Figure 2 demonstrate that persistent RNA·DNA hybrids readily form on GAA·TTC tracts during transcription in bacteria. The quantity of relaxed conformers formed after induction of transcription in bacteria (second to the last lane in Figure 2) rivals that attained during in vitro transcription. The resolution of relaxed conformers in the last lane by RNase H treatment verifies the RNA·DNA hybrid. The degree to which RNA·DNA hybrids are evident on the templates is particularly striking, because the bacterial strain used is wild type for RNase H, suggesting that the hybrids are formed more rapidly than they can be removed during active transcription of these plasmid templates.
The RNA·DNA hybrid can span the GAA·TTC repeat
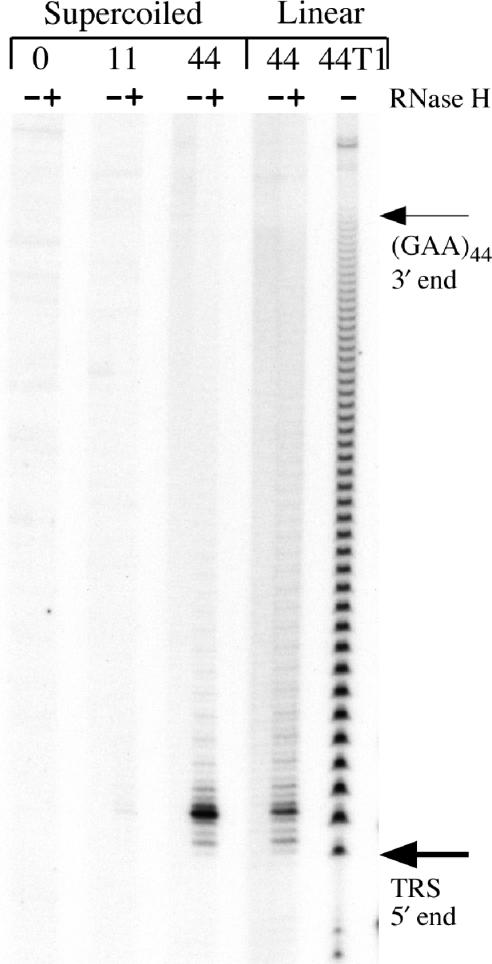
A sensitive estimate of the propensity for an RNA·DNA hybrid to form even transiently on a template can be provided by following the products of transcription coupled with RNase H digestion. T7 RNA polymerase initiates transcription with a guanine, so inclusion of gamma-labeled GTP in a transcription reaction allows for the 5′ end-labeling of transcripts during transcription, without post-processing.
RNase H digestion coupled to end-labeled transcription (Figure 3) confirms the length dependence of RNA·DNA hybrid formation within the GAA·TTC tract seen in the agarose gels of Figure 1. Only the template containing 44 triplets exhibits much RNase H sensitive material in Figure 3 (lanes 6 and 8). As expected, formation of the RNA·DNA hybrid target for RNase H is encouraged by pre-existing negative supercoils in the template. However, pre-existing negative supercoils are not required for formation of rGAA·dTTC hybrids as these also form on linear templates with a sufficiently long repeat tract (lane 8).
Figure 3.
The RNA·DNA hybrid extends to the promoter proximal end of the repeat tract. Pairs of transcription reactions end-labeled by including gamma 32P-GTP, were done in the absence (−) or presence (+) of RNase H to map the 5′ end of the RNA hybrid. Lanes 1–6 contain the end-labeled products derived from supercoiled templates with 0 (lanes 1 and 2), 11 (lanes 3 and 4) or 44 (lanes 5 and 6) GAA·TTC triplets. Lanes 7 and 8 contain the end-labeled products derived from a linear (GAA·TTC)44 template. The most common end-labeled fragments generated by RNase H were at or near the start of the GAA tract in the templates with 44 triplets (lanes 6 and 12). Lane 13 contains a ‘G ladder’ from a partial RNase T1 digest of end-labeled (GAA)44 transcript. The 3′ end of the (GAA)44 tract at base 187 is indicated by a thin arrow, the first G of the triplet repeat sequence, at base 55 in the transcript 5′ end is indicated by a thick arrow near the bottom of the gel. Full-length transcripts are not resolved on this gel (but see Figure 1).
The digestion of end-labeled reactions by RNase H also provides a way to map the 5′ limit of the hybrid within the transcript. In order to prepare an accurate RNA sizing ladder, we carefully titrated a (GAA)44 transcript with RNase T1, which specifically cleaves 3′ of single-stranded G residues. The lane labeled T1 in Figure 3 contains a partial digest of a 5′ end-labeled (GAA)44 transcript by RNase T1. Comparing the mobility of the major RNase H product in lanes 6 and 8 to the partial digest indicates that the 5′ end of the RNA in the hybrid maps very close to the 5′ end of the GAA tract in the transcript at base 55 (indicated by an arrow).
Transcription pauses are tightly linked to RNA·DNA hybrid formation
An extended RNA·DNA hybrid has been proposed to contribute to the arrest of eukaryotic RNA polymerase II in vitro (36), and RNA·DNA hybrids have been linked to transcription elongation deficits in yeast (16,17). We have previously demonstrated that transcribing T7 RNA polymerase will arrest in the distal part of a (GAA·TTC)88 repeat (28). We therefore investigated transcription and concomitant RNase H digestion of GAA·TTC bearing templates at high resolution, to determine whether RNA·DNA hybrid formation correlated with polymerase arrest.
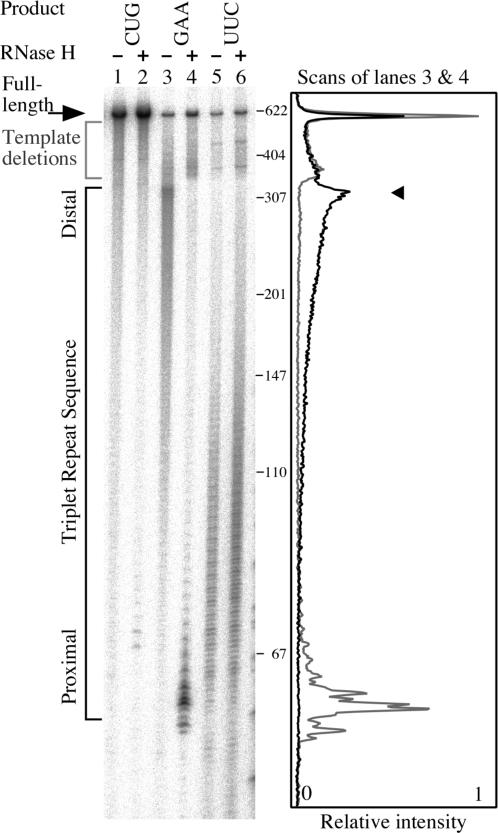
We made plasmids containing trinucleotide tracts of 88 repeats between a T7 promoter and a self-cleaving ribozyme, so that defined transcripts could be obtained from a supercoiled template (28). The pre-existing negative supercoils should encourage both structure formation by the template and RNA·DNA hybrid formation. We compared transcription in either direction through the GAA·TTC repeat, with and without the inclusion of RNase H. We performed this analysis on another disease causing triplet repeat, CTG·CAG, in parallel. The results of experiments like that shown in Figure 4, indicate that transcription arrest in the TTC repeat template is associated with RNA·DNA hybrid formation.
Figure 4.
RNA·DNA hybrids are associated with transcription arrest on TTC templates in vitro. Pairs of transcription reactions were done in the absence (−) or presence (+) of RNase H on supercoiled templates containing the triplet repeat sequences (CTG·CAG)88 (lanes 1 and 2), (GAA·TTC)88 (lanes 3 and 4) and (TTC·GAA)88 (lanes 5 and 6). The gel image shows the products of those reactions. To the right of the gel image the length in bases of select DNA size markers are indicated. At the far right of the figure is a scan of lanes 3 (dark line) and 4 (gray line) which highlights the RNase H mediated shift in truncation points from promoter distal (arrowhead) to promoter proximal within the GAA repeat. All the templates contain the sequence for a self-cleaving ribozyme that cuts the transcript 270 bases 3′ to the end of the repeat tract producing a full-length transcript of 590 bases. Transcription from the phage T7 promoter started 55 bases 5′ to the GAA·TTC insert. Transcripts were end-labeled by including gamma 32P-GTP in the reaction.
In lanes 1 and 2 of Figure 4, the results obtained with a (CTG·CAG)88 template show that neither transcription arrest nor RNase H sensitive RNA·DNA hybrid formation is a general feature of triplet repeats. Although minor stalling in the repeat is evident in lane 1, and faint product bands are visible in the sample treated with RNase H in lane 2, the degree to which these bands occur pales in comparison to those obtained with the (GAA·TTC)88 template.
When the GAA·TTC template is transcribed in the physiological direction (TTC template), the effect of RNase H digestion is striking. T7 RNAP is paused with increasing frequency towards the distal end of the repeat (Figure 4, lane 3). RNase H treatment completely shifts the pattern of pause sites from the distal half of the GAA·TTC repeat, to sharp peaks in the promoter proximal repeat (compare lanes 3 and 4 in Figure 4). The shift suggests that RNA hybrid formation in the entire repeat is tightly coupled to T7 RNAP arrest in the distal half of the repeat. RNase H treatment did not appreciably alter the pattern of stops that occurred outside the GAA·TTC repeat.
The GAA·TTC repeat can impede transcription from either direction. When transcription produces a UUC product, transcripts are frequently truncated in the promoter proximal half of the repeat (31). However, RNase H does not affect the intensity or distribution of truncation products from this direction of transcription in the GAA·TTC repeat (compare lanes 5 and 6 in Figure 4).
In yeast, RNA·DNA hybrid formation can mediate impaired transcription elongation at some distance downstream, due to tethering of RNA polymerase to the hybrid (17). A self-cleaving ribozyme downstream of the hybrid can free the polymerase from the tether (17). We were interested in whether a hybrid could form in the GAA·TTC repeat without causing immediate arrest within the repeat. We performed a variation on the experiment shown in Figure 4, in order to focus on polymerase that had made it beyond the repeat. In these experiments, unlabeled transcription reactions were performed in the absence or presence of RNase H. RNA purified from these reactions was then used as a substrate for primer extension by reverse transcriptase.
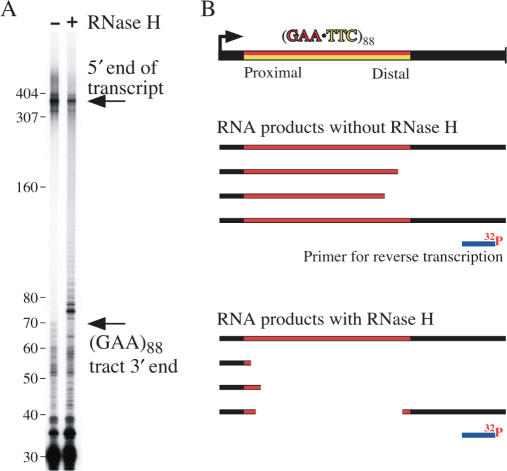
The results presented in Figure 5 show that if transcription is performed in the presence of RNase H, much of the primer extension product terminates within the repeat. Stops in the RNA template 3′ to the repeat are similar in both lanes. The GAA portion of the RNA apparently provides a good template for reverse transcriptase, as there is little stopping evident within the repeat in the absence of RNase H treatment. The extended product from the primer reached the 3′ end of the repeat tract at 72 bases (arrow) and the 5′ end of the transcript at 390 bases (near the top of the gel). The major RNase H specific bands migrate close to the end of the TRS. Thus, the transcripts those products represent had an RNA·DNA hybrid within the repeat but extended at least 72 bases beyond the repeat tract. The RNA·DNA hybrid did not extend beyond the repeat to the primer-binding site in the species represented by truncations within the repeat in Figure 5. We cannot estimate the degree to which RNA·DNA hybrids were continuous through the repeat and into the flanking sequence, since those would be completely digested with RNase H, and not bind the primer.
Figure 5.
A class of transcripts cleaved by RNase H can extend beyond the GAA·TTC repeat. (A) Primer extension provides high resolution mapping of the 3′ limit of the RNA·DNA hybrid within the transcript. The supercoiled (GAA·TTC)88 template used in Figure 4 was transcribed in the presence or absence of RNase H to provide RNA for reverse transcriptase primer extension of an end-labeled 30 base oligonucleotide that annealed from 72 to 42 bases beyond the 3′ end of the repeat in the transcript. Full-length extension to the 5′ end of the transcript yields a 390 base product in both samples. The major RNase H cleavage endpoints are clustered within a couple of triplets from the promoter distal end of the repeat sequence (arrow). (B) Experimental design and interpretation.
DISCUSSION
We have shown that stable RNA·DNA hybrids form readily during transcription of GAA·TTC repeats in vitro and in bacteria. The data indicate a strong propensity for RNA·DNA hybrid formation in the GAA·TTC repeat, link the hybrid to RNA polymerase arrest, and suggest that hybrid formation during transcription is intrinsic to long GAA·TTC repeats.
Other sequences that are purine-rich (especially G-rich) on the non-template strand have also been shown to have an intrinsic propensity to form RNA·DNA hybrids. For example, the non-template strands of the immunoglobulin class switch regions have a general purine bias featuring repeated sequences that are 60–70% purine, as well as regions of relatively uninterrupted purine runs (37). The immunoglobulin class switch regions have long been known to form RNA·DNA hybrids when transcribed in vitro. Moreover, RNA·DNA hybrids form only if the switch regions are transcribed in the physiological direction, suggesting a role for the hybrids in class switch recombination (20,21). Bisulfite modification of single-stranded cytosines displaced in the R-loop has been used to verify that RNA·DNA hybrids do indeed form during switch region transcription in activated B-cells (38). Thus, the propensity that the immunoglobulin class switch regions exhibit for RNA·DNA hybrid formation in vitro manages to overcome mechanisms that repress hybrid formation in the cell. While we would like to probe for RNA·DNA hybrid formation in the native FXN GAA·TTC repeat using bisulfite modification, it is not possible due to the lack of cytosines on the non-template strand. Moreover, while RNA·DNA hybrids have been found to extend to some degree both upstream and downstream of the purine-rich immunoglobulin switch region repeats (39), we have found that the ends of the GAA·TTC repeat act as a boundary to the hybrid. Consequently, bisulfite modification of DNA flanking the FXN repeat is not likely to be informative.
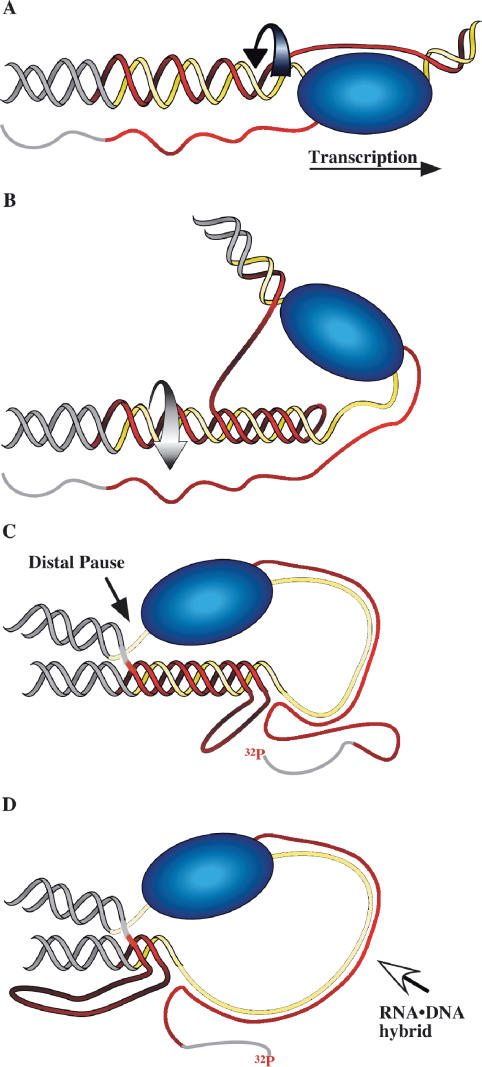
RNA·DNA hybrid formation adds to the already extensive repertoire of non-B DNA structures that can be adopted by GAA·TTC repeats. Like other uninterrupted purine·pyrimidine (R·Y) sequences, the repeat can form intermolecular and intramolecular DNA triple helices of either the R·R·Y or Y·R·Y configuration (40). An attractive hypothesis for RNA·DNA hybrid formation is a transient single-stranded state for the template strand when the non-template strand is engaged as the third strand in a triplex (20,21,41). Our working model, shown in schematic form in Figure 6, assumes that the transcript exits RNAP through the RNA exit pore in the normal way. We posit that negative supercoiling behind the advancing polymerase prompts formation of a short-lived intramolecular R·R·Y triplex in the GAA·TTC tract. This leaves part of the TTC template strand temporarily single stranded. The transcript would then simply re-anneal to the template strand after exiting RNA polymerase, with lessened competition from the non-template strand. Once annealed to the single-stranded region, the transcript can further displace the non-template strand, as shown in panel D of Figure 6. We have no evidence that the GAA transcript acts as a third strand in a triplex, nor do we think that it invades the duplex DNA independent of the transcribing polymerase. Consequently, while chromatin should inhibit access to the DNA by an exogenous complementary RNA, co-transcriptional RNA·DNA hybrid formation may bypass such inhibition because RNA polymerase has locally dislodged the histones from the template.
Figure 6.
Transcription-coupled RNA·DNA hybrid formation in a GAA·TTC repeat. Model for transient transcription-dependent triplex formation leading to an RNA polymerase pause and RNA·DNA hybrid formation. The purine (GAA or R) strand of the repeat is red, the pyrimidine (TTC or Y) strand is yellow and the flanking DNA is gray. (A) A standing wave of negative supercoiling follows RNA polymerase. At the transcription bubble, the non-template (GAA) strand is available to fold back in an R·R·Y interaction; the template strand is covered by RNA polymerase. (B) Rotation of the helix (curved arrow) as it winds in the third strand relaxes the negative supercoils caused by transcription and leaves a length of the template single-stranded. (C) RNAP is impeded at the distal template–triplex junction and the nascent transcript can anneal to the single-stranded stretch of template. (D) The RNA·DNA hybrid displaces the much less stable triplex structure. Structures of this type can account for the data generated by the 32P-end-labeled transcripts shown in lanes 3 and 4 of Figure 4.
An alternative model for reduced non-template competition suggested for immunoglobulin RNA·DNA hybrid formation is formation of G-quartet structures by the G-rich non-template strand (42). However, G-quartets are unlikely to contribute to hybrid formation in the GAA·TTC repeat. Occupation of the non-template strand by hairpin structures might also be expected to enhance hybrid formation, and CAG·CTG repeats readily form hairpins (43). We found that transcription through supercoiled CAG·CTG templates did not lead to extensive RNA·DNA hybrid formation. Thus, RNA·DNA hybrid formation in the FRDA repeat is not simply the consequence of potential structure formation by the non-template strand or the enhanced ability of simple repeat transcripts to re-anneal to the template at almost any point of contact.
RNA·DNA hybrid formation is likely to be a combination of several factors. Structure formation by the non-template strand may increase opportunities for the transcript to anneal. The lack of complexity in the repetitive sequence makes annealing simple. Indeed, it is possible that misaligned pairing of the rGAA transcript to the dTTC template contributes to the lack of branch migration beyond the confines of the repeat tract. Finally, the enhanced hybrid strength of rG·dC pairings and the absence of the weaker, disrupting rU·dA hybrids contribute to the stability of the rGAA·dTTC hybrid (44–46). The importance of a purine-rich transcript to hybrid stability is clear when considering transcription of the GAA·TTC repeat in the non-physiological direction. The GAA·TTC repeat has been shown by multiple groups to adopt a stable intramolecular Y·R·Y triplex that leaves a portion of the GAA strand unpaired (28,47,48). When transcribed to make CUU, the GAA·TTC repeat also forms an intramolecular acid stabilized Y·R·Y triplex capable of blocking transcription elongation (28,31). However, despite the single-stranded length of GAA template strand that is a consequence of an intramolecular triplex, we have found that the CUU transcript does not form stable, or even detectable, RNA·DNA hybrids. This is most likely due to the U-rich nature of the CUU transcript. The rU·dA base pairs are weaker than dT·dA base pairs because uracil lacks the C-5 methyl group of thymine, which is thought to contribute to base-stacking interactions (45).
In the physiological direction of transcription, we have shown that an extensive RNA·DNA hybrid is tightly linked to T7 RNA polymerase arrest in a TTC repeat template. In the nucleus, RNP formation should generally help block formation of RNA·DNA hybrids. However, in FRDA the GAA repeat within an FXN transcript numbers hundreds to thousands and it is possible that a protein with some specificity for rGAA is simply insufficient to bind it all. Huertas and Aguilera found that the selective transcription elongation impairment seen in mutants of the yeast THO/TREX complex (16) was due to RNA·DNA hybrid formation (17). Moreover, transcription elongation was improved in these yeast mutants if RNase H was over expressed, leading those authors to suggest that impaired elongation was likely due to the polymerase being tethered to an RNA·DNA hybrid (17).
The mechanism by which GAA·TTC expansions may impede transcription elongation in human cells is still unclear. Most models predict an obstruction to transcription elongation within the repeat, mediated either by DNA structure (25–27), or by altered chromatin (29,30). Our working model for transcription inhibition differs from most in that it predicts arrest downstream of a structure that includes an RNA·DNA hybrid (28,49). The results presented in Figure 4 lend further support for that model, and link the RNA·DNA hybrid to transcription arrest in vitro. On the other hand, experiments like that shown in Figure 5 suggest that transcription arrest is stochastic, as some transcripts extending beyond the repeat had formed a hybrid within the repeat.
Large GAA·TTC expansions in FRDA are unstable between generations (50,51) and show somatic mosaic expansion in individuals (52) implying a high degree of instability. While most long trinucleotide repeats are unstable to some degree, with mismatch repair implicated in their continued expansion with age (53,54), structure formation by GAA·TTC repeats may provide additional pathways to instability. We have shown that RNA·DNA hybrids form even on ‘pre-mutation’ size GAA·TTC repeats of about 40 triplets. This length does not cause FRDA disease symptoms, but is near the threshold of instability in both prokaryotic and eukaryotic systems. Given the recent linking of RNA·DNA hybrid formation to genome instability (16–19), it is plausible that RNA·DNA hybrid formation contributes to GAA·TTC repeat instability in the cell.
ACKNOWLEDGEMENTS
This research was supported by NIH grant number R01NS046567 from the National Institute Of Neurological Disorders and Stroke and by a grant from the Friedreich's Ataxia Research Alliance (FARA) to EG. Funding to pay the Open Access publication charges for this article was provided by grant R01NS046567.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sugino A, Hirose S, Okazaki R. RNA-linked nascent DNA fragments in Escherichia coli. Proc. Natl Acad. Sci. USA. 1972;69:1863–1867. doi: 10.1073/pnas.69.7.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang M, Ma N, Vassylyev DG, McAllister WT. RNA displacement and resolution of the transcription bubble during transcription by T7 RNA polymerase. Mol. Cell. 2004;15:777–788. doi: 10.1016/j.molcel.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 3.Datta K, Johnson NP, von Hippel PH. Mapping the conformation of the nucleic acid framework of the T7 RNA polymerase elongation complex in solution using low-energy CD and fluorescence spectroscopy. J. Mol. Biol. 2006;360:800–813. doi: 10.1016/j.jmb.2006.05.053. [DOI] [PubMed] [Google Scholar]
- 4.Naryshkina T, Kuznedelov K, Severinov K. The role of the largest RNA polymerase subunit lid element in preventing the formation of extended RNA-DNA hybrid. J. Mol. Biol. 2006;361:634–643. doi: 10.1016/j.jmb.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 5.Toulokhonov I, Landick R. The role of the lid element in transcription by E. coli RNA polymerase. J. Mol. Biol. 2006;361:644–658. doi: 10.1016/j.jmb.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 6.Crouch RJ. Ribonuclease H: from discovery to 3D structure. New Biol. 1990;2:771–777. [PubMed] [Google Scholar]
- 7.Wang JC. DNA Topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 8.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brill SJ, Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988;54:403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- 10.Makarova OV, Makarov EM, Sousa R, Dreyfus M. Transcribing of Escherichia coli genes with mutant T7 RNA polymerases: stability of lacZ mRNA inversely correlates with polymerase speed. Proc. Natl Acad. Sci. USA. 1995;92:12250–12254. doi: 10.1073/pnas.92.26.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masse E, Drolet M. Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J. Biol. Chem. 1999;274:16659–16664. doi: 10.1074/jbc.274.23.16659. [DOI] [PubMed] [Google Scholar]
- 12.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 13.Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurt E, Luo MJ, Rother S, Reed R, Strasser K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc. Natl Acad. Sci. USA. 2004;101:1858–1862. doi: 10.1073/pnas.0308663100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rondon AG, Jimeno S, Garcia-Rubio M, Aguilera A. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 2003;278:39037–39043. doi: 10.1074/jbc.M305718200. [DOI] [PubMed] [Google Scholar]
- 17.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Luna R, Jimeno S, Marin M, Huertas P, Garcia-Rubio M, Aguilera A. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol. Cell. 2005;18:711–722. doi: 10.1016/j.molcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Reaban ME, Griffin JA. Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature. 1990;348:342–344. doi: 10.1038/348342a0. [DOI] [PubMed] [Google Scholar]
- 21.Daniels GA, Lieber MR. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 1995;23:5006–5011. doi: 10.1093/nar/23.24.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 23.Filla A, De Michele G, Cavalcanti F, Pianese L, Monticelli A, Campanella G, Cocozza S. The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am. J. Hum. Genet. 1996;59:554–560. [PMC free article] [PubMed] [Google Scholar]
- 24.Montermini L, Richter A, Morgan K, Justice CM, Julien D, Castellotti B, Mercier J, Poirier J, Capozzoli F, et al. Phenotypic variability in Friedreich ataxia: role of the associated GAA triplet repeat expansion. Ann. Neurol. 1997;41:675–682. doi: 10.1002/ana.410410518. [DOI] [PubMed] [Google Scholar]
- 25.Bidichandani SI, Ashizawa T, Patel PI. The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am. J. Hum. Genet. 1998;62:111–121. doi: 10.1086/301680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohshima K, Montermini L, Wells RD, Pandolfo M. Inhibitory effects of expanded GAA.TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J. Biol. Chem. 1998;273:14588–14595. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- 27.Sakamoto N, Chastain PD, Parniewski P, Ohshima K, Pandolfo M, Griffith JD, Wells RD. Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich's ataxia. Mol. Cell. 1999;3:465–475. doi: 10.1016/s1097-2765(00)80474-8. [DOI] [PubMed] [Google Scholar]
- 28.Grabczyk E, Usdin K. The GAA·TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–913. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 30.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat. Chem. Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 31.Grabczyk E, Usdin K. Alleviating transcript insufficiency caused by Friedreich's ataxia triplet repeats. Nucleic Acids Res. 2000;28:4930–4937. doi: 10.1093/nar/28.24.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grabczyk E, Usdin K. Generation of microgram quantities of trinucleotide repeat tracts of defined length, interspersion pattern, and orientation. Anal. Biochem. 1999;267:241–243. doi: 10.1006/abio.1998.2962. [DOI] [PubMed] [Google Scholar]
- 33.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 35.Masse E, Drolet M. R-loop-dependent hypernegative supercoiling in Escherichia coli topA mutants preferentially occurs at low temperatures and correlates with growth inhibition. J. Mol. Biol. 1999;294:321–332. doi: 10.1006/jmbi.1999.3264. [DOI] [PubMed] [Google Scholar]
- 36.Kireeva ML, Komissarova N, Kashlev M. Overextended RNA:DNA hybrid as a negative regulator of RNA polymerase II processivity. J. Mol. Biol. 2000;299:325–335. doi: 10.1006/jmbi.2000.3755. [DOI] [PubMed] [Google Scholar]
- 37.Dunnick W, Hertz GZ, Scappino L, Gritzmacher C. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 1993;21:365–372. doi: 10.1093/nar/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 39.Huang FT, Yu K, Hsieh CL, Lieber MR. Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc. Natl Acad. Sci. USA. 2006;103:5030–5035. doi: 10.1073/pnas.0506548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frank-Kamenetskii MD, Mirkin SM. Triplex DNA structures. Annu. Rev. Biochem. 1995;64:65–95. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- 41.Grabczyk E, Fishman MC. A long purine-pyrimidine homopolymer acts as a transcriptional diode. J. Biol. Chem. 1995;270:1791–1797. doi: 10.1074/jbc.270.4.1791. [DOI] [PubMed] [Google Scholar]
- 42.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petruska J, Arnheim N, Goodman MF. Stability of intrastrand hairpin structures formed by the CAG/CTG class of DNA triplet repeats associated with neurological diseases. Nucleic Acids Res. 1996;24:1992–1998. doi: 10.1093/nar/24.11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ratmeyer L, Vinayak R, Zhong YY, Zon G, Wilson WD. Sequence specific thermodynamic and structural properties for DNA.RNA duplexes. Biochemistry. 1994;33:5298–5304. doi: 10.1021/bi00183a037. [DOI] [PubMed] [Google Scholar]
- 45.Wang S, Kool ET. Origins of the large differences in stability of DNA and RNA helices: C-5 methyl and 2′-hydroxyl effects. Biochemistry. 1995;34:4125–4132. doi: 10.1021/bi00012a031. [DOI] [PubMed] [Google Scholar]
- 46.Lesnik EA, Freier SM. Relative thermodynamic stability of DNA, RNA, and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry. 1995;34:10807–10815. doi: 10.1021/bi00034a013. [DOI] [PubMed] [Google Scholar]
- 47.Shimizu M, Hanvey JC, Wells RD. Intramolecular DNA triplexes in supercoiled plasmids. I. Effect of loop size on formation and stability. J. Biol. Chem. 1989;264:5944–5949. [PubMed] [Google Scholar]
- 48.Gacy AM, Goellner GM, Spiro C, Chen X, Gupta G, Bradbury EM, Dyer RB, Mikesell MJ, Yao JZ, et al. GAA instability in Friedreich's Ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol. Cell. 1998;1:583–593. doi: 10.1016/s1097-2765(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 49.Grabczyk E, Kumari D, Usdin K. Fragile X syndrome and Friedreich's ataxia: two different paradigms for repeat induced transcript insufficiency. Brain Res. Bull. 2001;56:367–373. doi: 10.1016/s0361-9230(01)00572-x. [DOI] [PubMed] [Google Scholar]
- 50.Monros E, Molto MD, Martinez F, Canizares J, Blanca J, Vilchez JJ, Prieto F, de Frutos R, Palau F. Phenotype correlation and intergenerational dynamics of the Friedreich ataxia GAA trinucleotide repeat. Am. J. Hum. Genet. 1997;61:101–110. doi: 10.1086/513887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Michele G, Cavalcanti F, Criscuolo C, Pianese L, Monticelli A, Filla A, Cocozza S. Parental gender, age at birth and expansion length influence GAA repeat intergenerational instability in the X25 gene: pedigree studies and analysis of sperm from patients with Friedreich's ataxia. Hum. Mol. Genet. 1998;7:1901–1906. doi: 10.1093/hmg/7.12.1901. [DOI] [PubMed] [Google Scholar]
- 52.Montermini L, Kish SJ, Jiralerspong S, Lamarche JB, Pandolfo M. Somatic mosaicism for Friedreich's ataxia GAA triplet repeat expansions in the central nervous system. Neurology. 1997;49:606–610. doi: 10.1212/wnl.49.2.606. [DOI] [PubMed] [Google Scholar]
- 53.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 54.van den Broek WJ, Nelen MR, Wansink DG, Coerwinkel MM, te Riele H, Groenen PJ, Wieringa B. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]