Abstract
There have been a growing number of studies where molecular beacons (MBs) are used to image RNA expression in living cells; however, the ability to make accurate measurements can be hampered by the generation of false-positive signals resulting from non-specific interactions and/or nuclease degradation. In the present study, we found that such non-specific signals only arise in the nucleus of living cells. When MBs are retained in the cytoplasmic compartment, by linking them to quantum dots (QDs), false-positive signals are reduced to marginal levels. Consequently, MB–QD conjugates were used to measure the expression of the endogenous proto-oncogene c-myc in MCF-7 breast cancer cells by quantifying the total fluorescent signal emanating from individual cells. Upon the addition of tamoxifen, measurements of MB fluorescence indicated a 71% reduction in c-myc expression, which correlated well with RT-PCR measurements. Variations in MB fluorescence resulting from instrumental fluctuations were accounted for by imaging fluorescent calibration standards on a daily basis. Further, it was established that measurements of the total fluorescent signal were not sensitive to the focal plane. Overall, these results provide evidence that accurate measurements of RNA levels can be made when MBs are retained in the cytoplasm.
Variations in gene expression are commonly considered the major determinant for dictating cell behavior. Accordingly, methods to measure gene expression, such as reverse-transcriptase (RT) PCR and DNA microarrays, have proven to be invaluable with regard to understanding cell regulatory processes and disease mechanisms. However, these methods generally provide only the relative change in gene expression for a population of cells. Under many circumstances it is the aberrant behavior of only a few cells or the stochasticity of RNA expression within a population that may be of interest (1–6).
In order to gain a better understanding of cell-to-cell variations in RNA expression, much effort has recently been devoted to developing methods for imaging RNA in single living cells. Currently, the majority of live cell imaging approaches utilize MBs, which are antisense oligonucleotide probes labeled with a ‘reporter’ fluorophore at one end and with a quencher at the other end (7). In the absence of complementary nucleic acid targets, the MBs assume a hairpin conformation, which brings the fluorophore and quencher into close proximity and creates a low-fluorescence or ‘dark’ state. Upon hybridization with complementary targets the fluorophore becomes separated from the quencher and fluorescence is restored. The unique ability of MBs to convert target recognition into a measurable fluorescent signal has led to their use in numerous applications ranging from monitoring the distribution and transport of β-actin mRNAs in motile fibroblasts to monitoring viral growth in living cells (8–18).
Despite their growing use, much debate remains over whether MBs emit false-positive signals due to non-specific interactions and/or degradation once introduced into living cells. This possibility is fueled by several studies where MBs were used to detect protein–DNA interactions and the presence of nucleases in solution (19,20). Further, there is evidence that MBs are quickly sequestered into the nucleus via facilitated transport, again suggesting undesirable protein–MB intracellular interactions in living cells (8). In fact, one study reported that linear nuclease resistant 2′-O-methyl probes performed just as well as 2′-O-methyl MBs in terms of RNA detection in living cells (21). It was hypothesized that this was due to opening of MBs by mechanisms other than hybridization.
In order to better understand whether MBs emit false-positive signals once introduced into living cells, we microinjected MBs that were not perfectly complementary to any known endogenous RNA into MCF-7 breast cancer cells. The total fluorescent signal emanating from single cells was then quantified over time. We observed a false-positive fluorescent signal within the nucleus, but not the cytoplasm. Once MBs were prevented from gaining entry into the nucleus by conjugating them to quantum dots (QDs), we no longer observed a non-specific fluorescent signal and were able to make sensitive measurements of endogenous gene expression. Specifically, we measured the expression of c-myc, an oncogene implicated in a variety of breast cancers (22–26). We also conducted experiments to evaluate the effect of tamoxifen, an anti-cancer drug, on the suppression of c-myc expression. Results were compared with quantitative RT-PCR.
METHODS
Molecular beacon design and synthesis
All oligonucleotide probes and targets were obtained from Integrated DNA Technologies, Inc. (Coralville, IA, USA). Antisense Firefly luciferase (pGL3-Luc 235-252, Promega) and antisense c-myc (564-581, GenBank Accession V00568) MBs were labeled at the 5′-end with a CAL Fluor® Red 610 (Biosearch Technologies) fluorophore, Cal610, and at the 3′-end with Iowa Black RQ quencher, IBRQ. Anti-luciferase MBs were used as a control sequence for these studies because this sequence is not complementary to any endogenous RNA target in MCF-7 cells. For studies requiring quantum-dot conjugations, a biotin-dT group was incorporated in the 3′-stem. The anti-luciferase MB employed was Cal610-GTCACCTCAGCGTAAGTGATGTCG(bio-dT)GAC-IBRQ. The c-myc antisense MB employed was Cal610-GTCACGTGAAGCTAACGTTGAGGG(bio-dT)GAC-IBRQ. Luciferase and c-myc target DNA oligonucleotides were synthesized with the sequences, GTCACGACATCACTTACGCTGAGTTT and GTCACCCTCAACGTTAGCTTCACTTT, respectively. An antisense c-myc 2′-O-methyl RNA oligonucleotide was synthesized with the sequence mGmUmGmAmAmGm CmUmAmAmCmGmUmUmGmAmGmG.
Synthesis and spectral analysis of quantum dot-molecular beacon conjugates
QD–MB conjugates (QD–MBs) were prepared using biotinylated MBs and QD streptavidin conjugates (QD800, Invitrogen). Specifically, 10 μM samples of the MB were incubated with 1 μM QD streptavidin conjugates at molar ratios of 5:1, 6:1, 10:1, 15:1, 20:1 and 100:1 in 50 mM sodium borate, 0.05% Tween, pH 8.3 at 4°C overnight. The specified molar ratios were obtained by adjusting the volumes of the MB and QD sample accordingly. QD–MBs were purified from unbound MBs by gel chromatography (Superdex, GE Healthcare). The concentration of the purified QD–MB was determined by measuring the absorbance of QD800 (ε405nm = 8 000 000 cm−1M−1) on a Cary100 spectrophotometer (Varian). Throughout the text the concentration of the QD will be used to represent the QD–MB concentration. QD–MB conjugates possess an average diameter of 15–21 nm (27).
To ensure that the QD–MB samples were pure from unbound MBs, an aliquot of the sample was centrifuged on a Microcon YM-50 filter (50 kDa MW cutoff, Millipore). The eluent was then incubated in the presence of complementary target and tested for the presence of MBs by performing fluorescence measurements on a SPEX FluoroMax-3 spectrofluorometer (Horiba Jobin Yvon). Specifically, the maximum fluorescence intensity of the hybridized MB (Cal610 Exc. = 590 nm, Emm. = 610 nm) in the eluent was compared to that of completely hybridized MBs in the stock QD–MB sample. Typically, the fluorescent emission contributed by unbound MBs accounted for less than 2% of the QD–MB emission at 610 nm.
The emission profile of each QD–MB conjugate was acquired on the FluoroMax-3 spectrofluorometer by setting the excitation wavelength to 590 nm and recording the emission from 600 to 825 nm. These experiments were carried out in 50 mM sodium borate, 100 mM NaCl, 0.05% Tween, pH 8.3 (QMB Buffer) using 10 nM QD–MBs in the presence or absence of 250 nM complementary target. It was found that for MB:QD molar ratios above 15:1 there was no significant improvement in the maximum fluorescence achieved in the presence of excess target. Therefore, a MB:QD molar ratio of 15:1 was used for all subsequent experiments. Further hybridization experiments were carried out with 10 nM of the 15:1 QD–MBs in the presence of 10–250 nM complementary target. The peak MB fluorescence of this sample reached a plateau at a target concentration of ∼120 nM. This suggests that all of the MBs were hybridized at this target concentration. To estimate the concentration of MBs that would be completely hybridized in the presence of 120 nM target, hybridization experiments were conducted with 60, 90 or 120 nM MBs in the presence of 10–250 nM complementary target. It was found that the peak MB fluorescence of the 90 nM sample reached a plateau at a target concentration of ∼120 nM. This suggests that the QD–MB sample contains ∼90 nM MB. Since, the QD concentration is 10 nM, it is estimated that there are ∼9 MBs per QD. Although this measurement only provides an estimate of the MB-to-QD ratio, this approach was preferred over spectrophotometric methods because the high absorbance of the QD at the peak MB absorbance wavelength introduced variability between readings.
In order to evaluate the affect of conjugating MBs to QDs on the rate of hybridization, a 100 μl sample of 6 μM luciferase target was added to 100 μl of 450 nM antisense luciferase MBs or 100 μl of 50 nM QD–MB conjugates (which is equivalent to 450 nM MBs). The samples were mixed and fluorescent measurements were taken immediately on a SPEX FluoroMax-3 spectrofluorometer (Horiba Jobin Yvon) at 25°C. All kinetics studies were conducted in QMB buffer.
Fluorometric analysis of MB- and QD–MB-protein interactions
MB and QD–MBs were incubated with single-stranded binding (SSB) protein and bovine serum albumin (BSA) to examine whether specific and non-specific protein interactions could be responsible for generating false-positive signals. Specifically, 100 μl of 90 nM MBs were mixed with 100 μl of 0.2 mg/ml BSA (2.99 μM) or with 100 μl of 720 nM SSB protein in QMB buffer. Control samples consisted of 100 μl of 90 nM MBs mixed with 100 μl of 6 μM complementary target or 100 μl of QMB buffer alone. Fluorescent measurements (excitation: 590 nm, emission: 610 nm) were taken on a SPEX FluoroMax-3 spectrofluorometer (Horiba Jobin Yvon) at 25°C immediately following mixing. Fluorescence was monitored over the course of 30 min at 1-min intervals. End point measurements were taken at 24 h. MB fluorescence was normalized to the MB signal in the presence of excess target. Analogous studies were carried our using 10 nM QD–MBs, which is equivalent to 90 nM MBs assuming a labeling ratio of nine MBs per QD. All experiments were carried out in triplicates.
The ability of various nucleases to degrade MBs and QD–MBs and generate false-positive signals was also examined. Specifically, 100 μl of 90 nM MBs were mixed with 100 μl of the 5′-exonuclease phosphodiesterase I (2.25 mU) or with 100 μl of the 3′-exonuclease phosphodiesterase II (2.25 mU) in QMB Buffer. Similarly, 100 μl of 90 nM MBs were mixed with 80 μl of Mung Bean endonuclease (2.24 mU) and 20 μl of 10× Mung Bean Buffer (Promega). Finally, 100 μl of 90 nM MBs were mixed with 80 μl of S1 endonuclease (2.24 mU) and 20 μl of 10× S1 nuclease buffer (Promega). Control samples consisted of 100 μl of 90 nM MBs mixed with 100 μl of 6 μM complementary target in each respective buffer or mixed with 100 μl of the buffer alone. Fluorescent measurements (excitation: 590 nm, emission: 610 nm) were taken on a SPEX FluoroMax-3 spectrofluorometer (Horiba Jobin Yvon) at 25°C immediately following mixing. Fluorescence was monitored over the course of 30 min at 1-min intervals. End point measurements were taken at 24 h. MB fluorescence was normalized to the MB signal in the presence of excess target in the respective buffer. Analogous studies were carried our using 10 nM QD–MBs. All experiments were carried out in triplicates.
Cell culture
Estrogen-responsive MCF-7 cells (ATCC, Manassas, VA, USA) were cultured in minimum essential medium (Eagle) with 2 mM l-glutamine and Earle's BSS adjusted to contain 1.5 g/l sodium bicarbonate, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate and 10% fetal bovine serum (FBS) in 5% CO2 at 37°C. It should be noted that estrogen was not removed from the FBS. Also, phenol red was present. It has been reported that phenol red can mimic the action of estradiol at a typical concentration of 30 μM (28). For all live cell-imaging experiments (described subsequently), the MCF-7 cells were seeded on a 60 mm petri dish at a confluency of 10–30%. In experiments where the effect of tamoxifen on c-myc expression was evaluated, MCF-7 cells were treated with 1 μM tamoxifen for 48 h prior to microinjection and imaging of QD–MBs. All cell experiments were carried out on cells passaged less than 20 times to ensure that they do not acquire resistance to tamoxifen (29).
Fluorescence microscopy
All microscopy measurements were performed on an Olympus IX 81 motorized inverted fluorescence microscope equipped with a Sensicam (Cooke) monochrome digital camera, an X-Cite 120 excitation source (EXFO) and Sutter excitation and emission filter wheels. Automated image acquisition was carried out using IPLab (BD Biosciences). Images of Cal610 and QD800 were acquired using the filter sets (HQ560/55, HQ645/75, Q595LP) and (e460spuv, D800/50, 475dcxru) (Chroma), respectively. A LUC PLAN FLN 40× objective (NA 0.6) was used for all cell imaging studies.
Water-in-oil bubble measurements
Fluorescent standards for comparing day-to-day fluctuations in the fluorescent intensity of the microscope set-up were generated by microinjecting samples containing 100 nM MBs and 1 μM complementary oligonucleotide targets in QMB buffer into paraffin oil. The MBs were hybridized to their complementary targets for 24 h prior to the first day of microinjections. The same sample was used for all fluorescent measurements over the course of the study. Fluorescent images of the water-in-oil bubbles were acquired using the Cal610 filter set. These images were subsequently analyzed with NIH Image J. Specifically, a region of interest (ROI) was drawn around each bubble and the total fluorescent intensity was measured. Similarly, the total fluorescence intensity from an equal sized ROI that was drawn around a ‘background’ region was also measured. The background subtracted fluorescence measurement for each bubble was then calculated, FMB. Measuring the diameter of the bubble and assuming a spherical geometry allowed the volume, V, of each bubble to be calculated. The value FMB/V was used to compare instrumental fluctuations in fluorescence intensity.
Water-in-oil microinjections studies were also conducted with 25 nM QD–MB conjugates (in the presence and absence of 2.5 μM targets). When QDs were injected into paraffin oil, there was no detectable signal in the Cal610 channel indicating no spectral overlap between the two fluorescent reporters. This is in agreement with measurements made on the spectrofluorometer. It should also be noted that fluorescence measurements were not very sensitive to the focal plane in which the images were acquired as long as an appropriate ROI encircling all of the fluorescence was utilized.
Single-cell fluorescence imaging and analysis
Microinjection
Microinjection of MCF-7 cells was carried out using a FemtoJet and Injectman NI 2 (Eppendorf) microinjection system fitted with Femtotips I (Eppendorf). Prior to use, Femtotips were treated with Hexamethyldisilazane (Fluka) for 10 min, followed by repeated washes in QMB buffer.
Image analysis
All image analyses were performed using NIH Image J. Specifically, an ROI was drawn around the injected cell and the total fluorescent intensity within the ROI was measured. In cells injected with QDs and QD–MB conjugates, the QD signal was used to help locate the cell. Background measurements were acquired by drawing ROIs outside the boundaries of the cell. The background subtracted fluorescent intensity from each MB image, FMB, and each QD image, FQD, was then calculated. All cells were analyzed as described here unless otherwise noted.
Focal plane measurements
Samples containing 100 nM QDs in QMB buffer were microinjected into MCF-7 cells. The cells were brought into focus, and then images were acquired from −21 μm below the focal plane to 15 μm above the focal plane in increments of 3 μm. Total cellular fluorescence was then determined for each image as described above.
Kinetic imaging
Samples containing 2.5 μM antisense luciferase MBs in QMB Buffer were microinjected into MCF-7 cells and imaged every 40 s for 46 min (n = 5). Samples containing 10 μM antisense luciferase MBs in QMB Buffer were microinjected into MCF-7 cells and imaged every 10 s for 100 s (n = 8). Competitive inhibitor studies were carried out by microinjecting MCF-7 cells with samples containing both 2.5 μM antisense luciferase MBs and 25 μM linear 2′-O-methyl RNAs targeting the same sequence (n = 4). The images within each time series were analyzed by drawing ROIs in the nucleus, cytoplasm and just outside the cell border (i.e. background). The same ROIs were used for all of the images in a particular time series. The mean intensity within the ‘background’ ROI was subtracted from mean intensity of the nucleus and cytoplasmic ROI and the resulting intensity values were normalized by the maximum nuclear fluorescence and plotted versus time for comparison. All kinetic measurements were performed at 25°C.
Ratiometric imaging in living cells
Antisense luciferase QD–MB conjugates at a concentration of 100 nM in QMB Buffer were either hybridized to excess oligonucleotide targets (2.5 μM) for 4 h or left unhybridized prior to microinjection into MCF-7 cells. Cells that were injected with QD–MBs in the absence of target (n = 6) were imaged every 5 min for 25 min to examine whether any noticeable degradation/non-specific interactions occurred, as potentially indicated by an increase in MB fluorescence. Fluorescence image acquisition and analysis was thenperformed as described above. Ratiometric measurements were calculated by simply dividing FMB by FQD. Cells that were injected with QD–MBs in the presence of target (n = 13) were imaged within 5 min following microinjection.
Imaging c-myc RNA expression
A 25 nM c-myc antisense QD–MB conjugates in QMB buffer were microinjected into untreated and tamoxifen-treated MCF-7 cells. Fluorescence image acquisition was then performed as described above. Images acquired ∼10 min following injection were used for analysis. Studies designed to determine the specificity of MB hybridization were carried out by co-injecting MCF-7 cells with 25 nM c-myc antisense QD–MB conjugates in the presence of excess linear 2′-O-methyl RNAs targeting the same sequence. Statistics comparing FMB of each experiment were performed using a two-tailed Student's t-test. Significant difference was taken at P < 0.01.
Quantitative RT-PCR
RT-PCR standards
c-myc RNA standards were created by performing in vitro transcription driven by a T7-promoter proceeding the coding sequence of c-myc cDNA (Origene) using the RiboMAX™ large-scale RNA production system as per manufacturer's protocol. The resulting RNA was purified using lithium chloride precipitation followed by ethanol precipitation. Briefly, an appropriate volume of LiCl was added into the transcription-mix to achieve a final concentration of 1.5 M. The mixture was then incubated on ice, spun at 20 000 g in 4°C, and the supernatant was discarded. The pellet was further washed with ice-cold 70% ethanol, dried in a speedvac apparatus and re-suspended in ice-cold nuclease-free water (Ambion). Following additional washes with ice-cold 100% ethanol in the presence of 0.3M sodium acetate and 70% ethanol, the purified RNA was subsequently dried in a speedvac apparatus and then re-suspended in 140 μl of nuclease-free water (Ambion). The concentration was determined by measuring the absorbance at OD260 on a Cary100 spectrophotometer (Varian) and serial dilutions were prepared for RT-PCR.
Quantitative RT-PCR
Cytoplasmic RNA from MCF-7 cells was isolated using the HighPure RNA isolation kit (Roche) and subsequently reversed-transcribed to cDNA using High Capacity cDNA reverse transcriptase kit (Applied Biosystems) as per manufacturer's protocol. Standard curves for the absolute quantification of c-myc RNA were generated by first adding known amounts of serially diluted c-myc RNA to the cell lysates of MCF-7 cells, immediately prior to RNA purification. Samples containing no additional c-myc RNA were used to generate a baseline measurement following RT-PCR. Quantitative RT-PCR was performed on ABI PRISM 7300 Sequence detection system using FAM-labeled Taqman® primer sets for c-myc and β-actin, and the Taqman® universal PCR Master Mix (Applied Biosystems) according to the manufacturer's protocol.
RESULTS AND DISCUSSION
Molecular beacons elicit a non-specific signal in the nucleus of living cells
To investigate whether MBs composed of a nuclease-vulnerable DNA backbone elicited a false-positive signal in living cells, we microinjected MBs that were not perfectly complementary to any known endogenous RNA into MCF-7 breast cancer cells. It was hypothesized that these MBs would remain quenched unless opened due to non-specific interactions and/or nuclease degradation. Consequently, any increase in fluorescence would indicate a false-positive event.
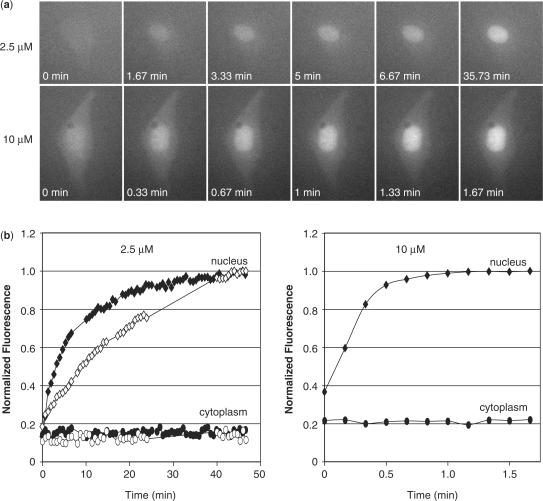
Immediately following the cytoplasmic microinjection of MBs, there was a clear increase in fluorescence in the nucleus, while there was no detectable change in fluorescence in the cytoplasm (Figure 1a). The fluorescent signal in the cytoplasm remained extremely faint and was barely distinguishable from the background. The rate of increase in fluorescence observed in the nucleus varied with injected dose, with lower concentrations of MBs taking longer before the fluorescent signal in the nucleus reached a plateau. Specifically, when MBs at a concentration of 10 μM were injected into the cytoplasm, the fluorescent signal within the nucleus typically reached a plateau between 1 and 2 min; however, when cells were injected with 2.5 μM of MBs the fluorescent signal did not plateau for 40–50 min (Figure 1b). This difference may largely be due to a faster diffusion into the nucleus, although a higher concentration of MBs may also occupy a higher percentage of the protein machinery, and thus expedite the process of active transport as well.
Figure 1.
Nuclear localization of MBs. (a) Fluorescent microscopy images of MCF-7 cells microinjected with either 2.5 μM or 10 μM samples of MBs. The MBs are not perfectly complimentary to any known endogenous RNA in MCF-7 cells. An increase in fluorescence was observed in the nucleus shortly after injection in all cells analyzed. (b) The mean fluorescent intensity measured in the nucleus (filled diamonds) and cytoplasm (filled circles) was plotted as a function time. The fluorescent signal increased in the nucleus but not the cytoplasm. The rate of increase depended on the injected concentration of MBs. Higher concentrations of MBs led to a more rapid nuclear localization. When MBs were co-injected with 2′-O-methyl RNAs with the same targeting-sequence into MCF-7 cells, there was still an increase in nuclear fluorescence (open diamonds); however, the rate of increase in fluorescence was slightly slower. The fluorescent signal within the cytoplasm remained low (open circles).
In order to determine whether the increase in fluorescence in the nucleus was in response to undesirable MB hybridization or due to non-specific protein interactions (or nuclease degradation), live cell competitive inhibition studies were performed. Specifically, an excess of linear 2′-O-methyl RNAs with the same targeting-sequence as the MBs were co-injected with MBs into MCF-7 breast cancer cells. It was expected that if the MB signal were due to target hybridization, then the 2′-O-methyl RNAs would compete for the same binding sites and reduce/eliminate the fluorescent signal. Despite injections of 10-fold excess 2′-O-methyl RNAs, we continued to observe an increase in MB fluorescence in the nucleus; however, the rate of increase was slightly reduced (Figure 1b). Presumably, a slower increase in fluorescence is due to the DNA MBs and the 2′-O-methyl RNAs utilizing some of the same machinery for active transport into the nucleus. Nonetheless, these observations suggest that the MB signal in the nucleus is likely to be non-specific, involving mechanisms other than hybridization.
MBs retained in the cytoplasm do not elicit false-positive signals
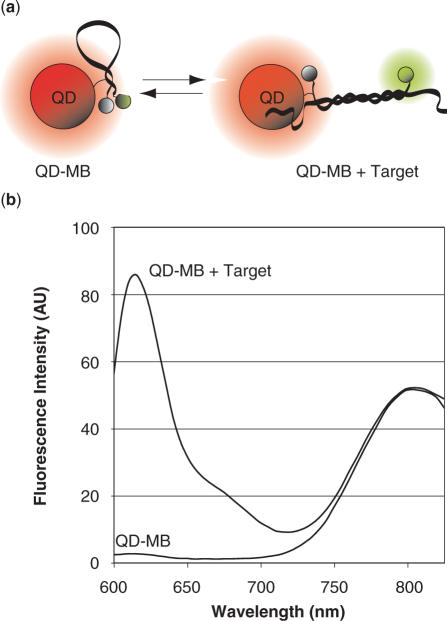
As noted earlier, when MBs with no known intracellular RNA target were injected into MCF-7 breast cancer cells only very faint fluorescent signals were observed in the cytoplasm. To investigate whether this was due to complete sequestration of the MBs into the nucleus or because MBs remained in a quenched hairpin conformation in the cytoplasm, we attached MBs to QDs so they would not be able to pass through the nuclear pores (Figure 2a).
Figure 2.
Schematic and emission spectrum of QD–MB conjugates. (a) Schematic of MB-QD conjugates in the presence and absence of target RNA. (b) The emission spectrum for 10 nM QMBs (Cal610 reporter, Iowa Black RQ quencher, and a QD800 quantum dot) was recorded in the absence and presence of excess complementary oligonucleotide targets. The addition of complementary target led to a large increase in MB fluorescence; however, the QD fluorescence remained unchanged. This labeling ratio (prior to purification) was 6 MBs per QD for the QD–MB emission spectrum shown.
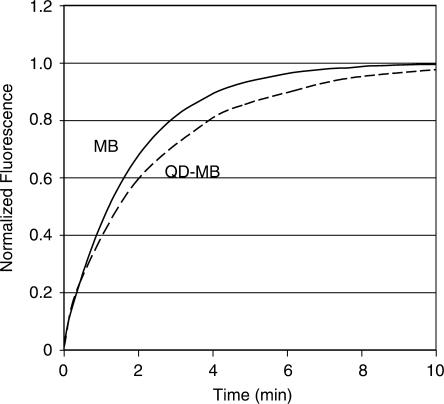
QDs were chosen over large non-karyophilic proteins such as streptavidin as a means of inhibiting entry into the nucleus because QDs also provide a bright and stable fluorescent signal that can be used for tracking MB localization. Further, the QD fluorescence allows for accurate ratiometric measurements since the QD signal remains unquenched regardless of the conformation of the probe (Figure 2b). Ratiometric measurements are helpful in removing experimental variability in single-cell kinetic studies because while changes in fluorescent intensity due to instrumental fluctuations will directly alter the intensity of the MB and QD signal individually, their ratio will remain unchanged. Of course, one concern with using QDs is that their large size might interfere with MB-target hybridization; however, comparison of the kinetics of in vitro hybridization of free MBs and MB-QD conjugates indicated that the QDs had relatively little affect on hybridization (Figure 3). This result is in agreement with previous studies where DNA attached to submicron latex particles exhibited rate constants similar (0.2× to 1×) to those of unconjugated DNA in homogeneous solutions (30). Since the diffusion boundary layer is so small on micro- and nanoparticles, slower hybridization kinetics would have likely meant that the QD–MB probe was reaction-rate limited (31). Perhaps, the separation between the MBs on each QD, likely due to the spacing of biotin binding domains, helps limit any type of steric interference during QD–MB-target hybridization.
Figure 3.
Hybridization kinetics. The rate of hybridization of free MBs (solid line) to complementary target and QD–MBs (dashed line) to complementary target were compared. Normalized fluorescent intensity is plotted versus time.
It has recently been reported that conjugation of MBs to QDs could result in fluorescence resonance energy transfer (FRET) (27). Indeed some FRET was observed with the QD–MB conjugates reported here. Specifically, the fluorescence of unhybridized MBs was quenched by 70% upon conjugation to QDs; however, upon the addition of excess complementary target the conjugated MBs only exhibited a 34% reduction in peak fluorescence compared with an equimolar amount of unconjugated MBs. These measurements assume a labeling ratio of nine MBs to each QD. The larger extent of quenching of MB-QD conjugates in the hairpin conformation leads to an improvement in signal-to-background (S:B = 93) compared with unconjugated MBs (S:B = 43). In contrast to the MB fluorescence, the QD signal is increased by 2.3% upon conjugation to MBs.
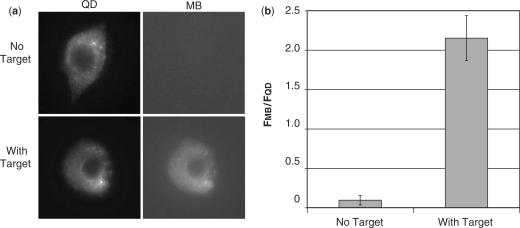
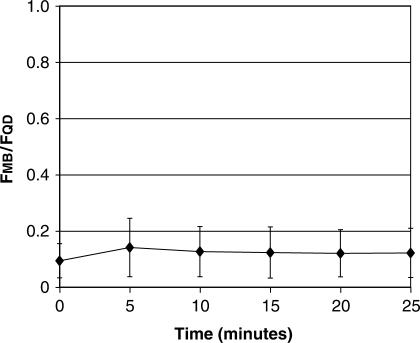
When non-targeted QD–MB conjugates were microinjected into living cells, the QD–MBs were localized entirely in the cytoplasmic compartment and no MB fluorescence was observed in the cell immediately after injection (Figure 4a), although image analysis did indicate an MB signal slightly above the background in all of the images. The average single-cell fluorescent ratio FMB/FQD was calculated to be 0.094 ± 0.061. No further increase in signal was observed for at least the next 20 min (Figure 5), indicating the absence of false-positive signals.
Figure 4.
Single cell measurements of MB and QD fluorescence. (a) QD–MBs in the absence of target or pre-hybridized to target we microinjected into MCF-7 cells. QD–MBs in the absence of target exhibited a bright fluorescent signal in the QD image but no signal was visible in the MB image. QD–MBs prehybridized to target exhibited a bright fluorescent signal in both the QD and MB images. In all images, the QD–MBs were only observed in the cytoplasmic compartment. (b) Single-cell measurements of total integrated MB fluorescence divided by total integrated QD fluorescence, following microinjection of QD–MBs in the absence of target (n = 19) and QD–MBs prehybridized to target (n = 13).
Figure 5.
Temporal measurements of QD–MB fluorescence in single living cells. QD–MBs that are not perfectly complementary to any known endogenous RNA were microinjected into MCF-7 breast cancer cells (n = 6). The ratio of total integrated MB fluorescence divided by total integrated QD fluorescence was monitored over time. No increase in MB fluorescence was observed for 25 min.
When QD–MB conjugates were pre-hybridized to complementary oligonucleotide targets prior to microinjection into MCF-7 cells, a bright fluorescent signal in the cell cytoplasm was visible in both the MB and QD images. The fluorescent ratio FMB/FQD was found to be 2.15 ± 0.29, which indicates an average intracellular signal-to-background ratio (i.e. [FMB/FQD]With Target/[FMB/FQD]No Target) of 23:1 (Figure 4b). The difference between intracellular and solution measurements of signal-to-background is likely due to lower signal-to-noise in live cell studies. Nonetheless, these findings provide evidence that the MBs remain in their quenched hairpin conformation when limited to the cytoplasmic compartment.
MB-protein interactions and MB degradation
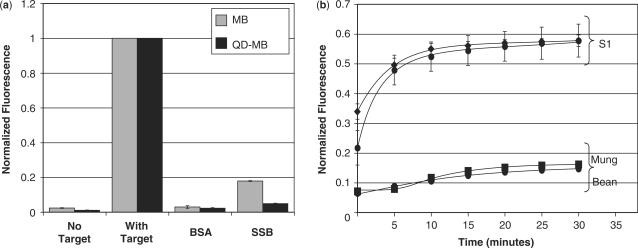
To gain insight into the cause of the false-positive signal observed in the nucleus following the microinjection of MBs into living cells and to determine the effect of conjugating MBs to QDs on these interactions, a fluorometric analysis of MBs and QD–MBs in the presence of various proteins and nucleuses was conducted. It was found that non-specific interactions with BSA did not result in a substantial increase in fluorescent emission for either the MBs or QD–MBs (Figure 6a); however, SSB protein did cause a significant increase in fluorescence. Specifically, the MB fluorescence increased to 17.9 ± 0.2% of the signal elicited by a completely hybridized MB, while the QD–MB fluorescence increased to 4.9 ± 0.2% of the signal elicited by a completely hybridized QD–MB. The lesser effect of SSBs on QD–MB fluorescence suggests that steric interference may limit SSB-MB binding when the MBs are conjugated to QDs.
Figure 6.
Fluorometric analysis of MB- and QD–MB-protein interactions and endonuclease degradation. (a) MBs and QD–MBs were incubated with molar excess of complementary target, BSA and SSB (n = 3 each) and the peak fluorescent intensity of the MB was measured. SSB generated a false-positive signal upon binding to MBs and QD–MBs, although the increase in signal was much less pronounced with QD–MBs. (b) Incubation of MBs and QD–MBs with S1 endonucleases resulted in a significant increase in fluorescence. Both probes were degraded at similar rates. Mung Bean nuclease was also capable of degrading MBs and QD–MBs but to a much lesser extent than S1 nucleases. All fluorescent measurements were normalized to the signal elicited by MBs and QD–MBs, respectively, in the presence of saturating concentrations of complementary target.
Previous studies have shown that when MBs (250 nM) are incubated in the presence of saturating concentrations of SSB (529 nM), the MB signal can increase to more than 90% of the signal elicited by a completely hybridized MB (19). It is hypothesized that the smaller increase observed in our experiments is due to incomplete binding at the lower concentrations of MB (45 nM) and SSB (360 nM) tested.
When MBs and QD–MBs were incubated with phosphodiesterase I (5′-exonuclease) and phosphodiesterase II (3′-endonuclease), only a small increase in MB and QD–MB fluorescence was observed over a period of 30 min (Supplementary Figure 1). Although a slightly faster rate of increase in fluorescence was observed with unconjugated MBs, the difference is negligible. It appears that the reporter fluorophore and quencher at the 5′- and 3′-end respectively limited exonuclease activity for both MBs and QD–MBs. In contrast to exonucleases, MBs and QD–MBs were both highly susceptible to degradation by S1 endonucleases (Figure 6b). The fluorescent signal of both probes increased to 60% of their maximum with 30 min. Mung Bean endonucleases, which are known to cleave hairpin loops, also degraded both MBs and QD–MBs, but to a much lesser extent than S1 nucleases. The rate of degradation was similar for MBs and QD–MBs in the presence of endonucleases.
Based on our findings, it appears that both single-stranded DNA binding proteins and endonucleases could be responsible for generating false-positive signals in the nucleus following the microinjection of MBs into living cells. Although SSB proteins and nucleases are also present in the cytoplasm, previous reports have demonstrated that there are fewer DNA–protein interactions in the cytoplasm compared with the nucleus and that nuclease activity in the cytoplasm is low (32,33). These differences could help explain why MBs only elicited detectable false-positive signals in the nucleus. Our findings also suggest that the QD–MB signal may remain quenched following microinjection into living cells not only because of cytoplasmic localization but also due to fewer interactions with SSB proteins.
Measurements of instrumental fluctuations in fluorescence
In general, when performing live cell studies with MBs it is expected that the total MB fluorescence within single cells would correlate directly with the copy number of target RNA. However, when comparing fluorescent images of cells that were acquired at different time points or on different days, the total fluorescent signal may not only reflect the extent of MB hybridization but also instrumental variability. For example, the intensity of MB fluorescence would scale directly with fluctuations in the intensity of the excitation light source. Although the effects of instrumental variability could potentially be removed with ratiometric imaging, this approach is only effective when monitoring fluorescence within a single cell. When mRNA levels in different cells are compared, ratiometric measurements would also be dependent on the efficiency of probe delivery, i.e. the number of QD–MBs delivered into a cell. For example, if twice the number of QD–MB conjugates are injected into a cell, then the fluorescent ratio would be reduced to half because the number of QDs would be doubled but the number of hybridized MBs (i.e. the number of RNA targets) would remain the same.
Since tightly controlling the number of probes delivered into a cell is very difficult, even with microinjection where cell volume, morphology, and the actual microinjection tip being used could all affect delivery, ratiometric imaging is unlikely to provide accurate measurements of gene expression. Therefore, we chose to image gene expression by directly quantifying the MB signal emitted by single cells. In order to reduce the impact of instrumental variability we developed a simple method to calibrate the fluorescent microscope each day or even multiple times a day so that absolute measurements of MB fluorescence could be acquired and the recorded values could be adjusted according to any instrumental fluctuations that were measured. It should be noted that absolute measurements of MB fluorescence are not as sensitive as ratiometric measurements to the injected dose of MBs because the fluorescent signal of the unhybridized MBs is quenched. Only at high concentrations (above ∼2 μM) was it possible to visualize the background fluorescence of quenched MBs above the noise.
The microscope calibration methodology involved injecting known concentrations of pre-hybridized MBs into paraffin oil, which results in the formation of spherical water-in-oil bubbles (Supplementary Figure 2 inset). With this approach, each bubble serves as a well-characterized fluorescent standard that can be imaged directly on the microscope. It was hypothesized that any changes that were observed in MB fluorescence would correspond to instrumental fluctuations. Although MBs were utilized here, they can easily be replaced with any fluorescent molecule.
Since the total fluorescence of each bubble increases linearly with volume, which can vary from bubble to bubble, fluorescent intensity measurements were compared only after being normalized by the respective bubble volume (FMB/V). These measurements were conducted multiple times a day for 1 week and whenever semi-quantitative measurements of fluorescence were being performed. We found that within any given day there were only very small fluctuations in the fluorescence intensity of the microscope, i.e an SD of less than 3% in fluorescent intensity measurements (Supplementary Figure 2); however, between days fluorescence intensity measurements differed by as much a 28%. This suggests that each time the light source is ignited it may have a slightly different intensity. Daily measurements of instrumental fluctuations in fluorescence intensity were used to adjust measurements of MB and QD fluorescence accordingly.
Focal plane has marginal effect on measurements of total cellular fluorescence
In addition to instrumental fluctuations, it may be speculated that another potential source of error when quantifying and comparing the fluorescence of multiple cells is any difference in the focal plane. When fluorescently labeled cells are observed under a microscope, there are clear changes in the peak fluorescent signal as the cells are brought in and out of focus. Cells that are in focus exhibit a very bright clear signal, while cells that are out of focus exhibit a fainter fluorescent signal that is diffuse over a much larger area. Despite these visual differences, when using wide-field fluorescent microscopy the number of fluorophores being excited and the number of photons being detected were hypothesized to be the same, assuming no major losses in light due to absorption between the cell and the objective. To validate that total cellular fluorescence is indeed independent of focal plane, QD–MB conjugates were microinjected into MCF-7 cells and images were acquired from 15 μm below the focal plan of the cell to 15 μm above the focal plane in increments of 3 μm (Supplementary Figure 3a). The total QD fluorescence in each image was then measured, making sure to include all of the fluorescent signal within the ROIs that were drawn around each cell. This often required drawing regions of interest well outside the cell itself. As shown in Supplementary Figure 3b, there appeared to be only a small loss in signal as the focal plane moved through the cells. In fact, the percent difference between the total fluorescent signal measured at the focal plane and the total fluorescent signal measured 15 μm above or below the focal plane was less than 3.3%. Therefore, it was concluded that quantitative measurements of QD and MB fluorescence are not dependent on the focal plane.
QD–MB conjugates allow for imaging of endogenous c-myc expression
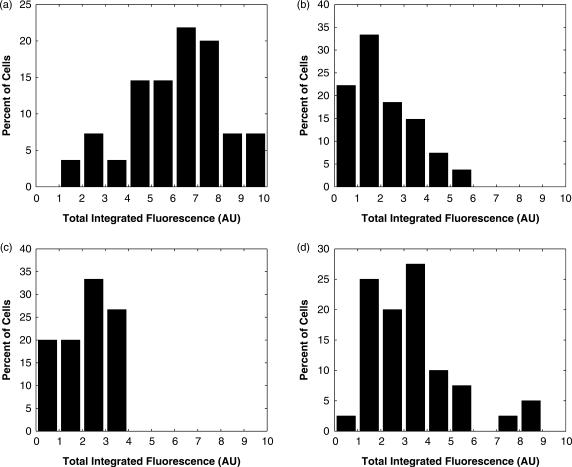
In order to evaluate whether QD–MB conjugates could be used to sensitively detect endogenous gene expression across a population of MCF-7 breast cancer cells, microinjection experiments were performed using antisense c-myc QD–MB conjugates. A fluorescent signal was detectable in the cytoplasm of all the microinjected cells. Interestingly, the difference in total integrated fluorescence between the faintest and brightest cell was more than 5-fold (Figure 7). It is hypothesized that this observed variation at least partly reflects the stochasticity in gene expression, which has been shown to play a pivotal role in governing cellular fates and disease evolution (1–6). However, instrumental noise also is surely a contributing factor.
Figure 7.
Measurements of MB fluorescence in MCF-7 cells. (a), MCF-7 cells were microinjected with antisense c-myc QD–MBs (n = 55) and the total integrated fluorescence within each cell in the MB images were determined. All measurements were normalized by the total fluorescence measured in the brightest cell and then multiplied by 10. (b) Total integrated MB fluorescence from MCF-7 cells co-microinjected with QD–MBs and 2′-O-methyl RNAs with the same targeting-sequence (n = 27). (c) Total integrated MB fluorescence from MCF-7 cells microinjected with MBs that are not perfectly complementary to any known endogenous RNA (n = 15). (d) Total integrated MB fluorescence from MCF-7 cells microinjected with antisense c-myc QD–MBs following treatment of cells with tamoxifen (n = 40).
Two negative control experiments were conducted to examine whether the measured fluorescent signal truly reflected MB hybridization or false-positive events. The first negative control consisted of injecting MBs with no known intracellular RNA target into MCF-7 cells. Image analysis of MB fluorescence revealed signals that were statistically lower (P < 0.01) than antisense c-myc signals. In general, the MB signal could not even be detected visually, and thus the QD fluorescence was necessary to identify the location of the cell. The second negative control consisted of competitively inhibiting MB hybridization with excess linear 2′-O-methyl RNAs targeting the same sequence. Again, a semi-quantitative analysis of MB fluorescence revealed a signal that was statistically lower than the antisense c-myc signals (P < 0.01). These results suggest that QD–MB conjugates can be used for the specific and sensitive detection of endogenous RNA in single living cells.
When MCF-7 cells were treated with tamoxifen, the average measure of total cellular fluorescence was 71% lower than in untreated MCF-7 cells. This is in agreement with previous reports, which found that tamoxifen caused a significant reduction in the global expression of c-myc (23,34,35). Interestingly, while the majority of the cells exhibited an extremely faint fluorescent signal similar to the negative controls, there were a small number of cells that exhibited a fluorescent signal similar to untreated cells. It is hypothesized that these outliers may represent a small percentage of cells that are resistant to tamoxifen.
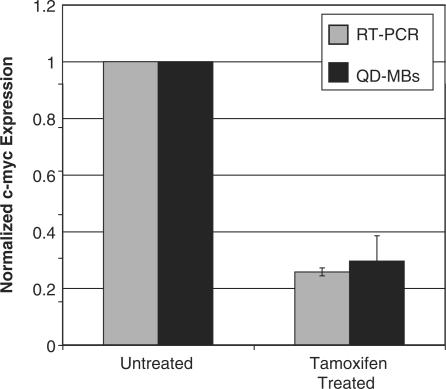
When Quantitative RT-PCR was carried out on MCF-7 cells with and without tamoxifen treatment, we observed a 74 ± 2% decrease in c-myc expression following tamoxifen treatment, which agrees well with the MB data (Figure 8). These results support the use of MBs that are restricted to the cytoplasmic compartment for the semi-quantitative measurement of endogenous gene expression. Quantitative RT-PCR revealed that the average copy number of c-myc mRNA per MCF-7 cell was 2907 ± 289 for untreated cells and 747 ± 101 for tamoxifen-treated cells. While this data provides insight into the sensitivity of QD–MB conjugates, it is important to note that absolute measurements of copy number are highly sensitive to RNA handling and methodology (36,37). For comparison, another report indicated a copy number of 8000 c-myc transcripts in MCF-7 cells (38,39).
Figure 8.
Effect of tamoxifen on c-myc expression. RT-PCR and QD–MBs both indicate a 70–75% downregulation in c-myc expression following treatment of MCF-7 cells with tamoxifen. The error bars represent the standard error.
In summary, we demonstrate that MBs only elicit a non-specific signal in the nucleus. When MBs are retained in the cytoplasmic compartment, by conjugation to QDs, false-positive signals are reduced to marginal levels. Variations in MB fluorescence resulting from instrumental fluctuations can be accounted for by imaging water-in-oil fluorescent calibration standards on a daily basis. As a result, QD–MB conjugates can be used as an effective tool for imaging the expression of specific mRNAs in single living cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This material is based upon work supported in part by the National Institute of Health (NCI) R21 CA116102, the National Science Foundation BES-0616031, and the American Cancer Society RSG-07-005-01. Funding to pay the Open Access publication charges for this article was also provided by the grants mentioned above.
Conflict of interest statement. I, Dr. Mark Behlke, am employed by Integrated DNA Technologies, Inc., (IDT) which offers oligonucleotides for sale similar to some of the compounds described in the manuscript. IDT is however not a publicly traded company and I personally do not own any shares/equity in IDT.
REFERENCES
- 1.McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc. Natl Acad. Sci. USA. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blake WJ, Kaern M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 3.Hume DA. Probability in transcriptional regulation and its implications for leukocyte differentiation and inducible gene expression. Blood. 2000;96:2323–2328. [PubMed] [Google Scholar]
- 4.Ross IL, Browne CM, Hume DA. Transcription of individual genes in eukaryotic cells occurs randomly and infrequently. Immunol. Cell. Biol. 1994;72:177–185. doi: 10.1038/icb.1994.26. [DOI] [PubMed] [Google Scholar]
- 5.Kepler TB, Elston TC. Stochasticity in transcriptional regulation: origins, consequences, and mathematical representations. Biophys. J. 2001;81:3116–3136. doi: 10.1016/S0006-3495(01)75949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl Acad. Sci. USA. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 8.Tyagi S, Alsmadi O. Imaging native beta-actin mRNA in motile fibroblasts. Biophys. J. 2004;87:4153–4162. doi: 10.1529/biophysj.104.045153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl Acad. Sci. USA. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santangelo PJ, Nix B, Tsourkas A, Bao G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perlette J, Tan W. Real-time monitoring of intracellular mRNA hybridization inside single living cells. Anal. Chem. 2001;73:5544–5550. doi: 10.1021/ac010633b. [DOI] [PubMed] [Google Scholar]
- 12.Nitin N, Santangelo PJ, Kim G, Nie S, Bao G. Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells. Nucleic Acids Res. 2004;32:e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mhlanga MM, Vargas DY, Fung CW, Kramer FR, Tyagi S. tRNA-linked molecular beacons for imaging mRNAs in the cytoplasm of living cells. Nucleic Acids Res. 2005;33:1902–1912. doi: 10.1093/nar/gki302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vargas DY, Raj A, Marras SA, Kramer FR, Tyagi S. Mechanism of mRNA transport in the nucleus. Proc. Natl Acad. Sci. USA. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santangelo P, Nitin N, LaConte L, Woolums A, Bao G. Live-cell characterization and analysis of a clinical isolate of bovine respiratory syncytial virus, using molecular beacons. J. Virol. 2006;80:682–688. doi: 10.1128/JVI.80.2.682-688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng XH, Cao ZH, Xia JT, Carlson GW, Lewis MM, Wood WC, Yang L. Real-time detection of gene expression in cancer cells using molecular beacon imaging: new strategies for cancer research. Cancer Res. 2005;65:1909–1917. doi: 10.1158/0008-5472.CAN-04-3196. [DOI] [PubMed] [Google Scholar]
- 17.Medley CD, Drake TJ, Tomasini JM, Rogers RJ, Tan W. Simultaneous monitoring of the expression of multiple genes inside of single breast carcinoma cells. Anal. Chem. 2005;77:4713–4718. doi: 10.1021/ac050881y. [DOI] [PubMed] [Google Scholar]
- 18.Drake TJ, Medley CD, Sen A, Rogers RJ, Tan W. Stochasticity of manganese superoxide dismutase mRNA expression in breast carcinoma cells by molecular beacon imaging. Chembiochem. 2005;6:2041–2047. doi: 10.1002/cbic.200500046. [DOI] [PubMed] [Google Scholar]
- 19.Li JJ, Fang X, Schuster SM, Tan W. Molecular beacons: a novel approach to detect protein - DNA interactions. This work was partially supported by a U.S. NSF Career Award (CHE-9733650) and by a U.S. Office of Naval Research Young Investigator Award (N00014-98-1-0621) Angew. Chem. Int. Ed. Engl. 2000;39:1049–1052. doi: 10.1002/(sici)1521-3773(20000317)39:6<1049::aid-anie1049>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Li JJ, Geyer R, Tan W. Using molecular beacons as a sensitive fluorescence assay for enzymatic cleavage of single-stranded DNA. Nucleic Acids Res. 2000;28:E52. doi: 10.1093/nar/28.11.e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molenaar C, Marras SA, Slats JC, Truffert JC, Lemaitre M, Raap AK, Dirks RW, Tanke HJ. Linear 2′ O-Methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res. 2001;29:E89–89. doi: 10.1093/nar/29.17.e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-Yosef T, Yanuka O, Halle D, Benvenisty N. Involvement of Myc targets in c-myc and N-myc induced human tumors. Oncogene. 1998;17:165–171. doi: 10.1038/sj.onc.1201939. [DOI] [PubMed] [Google Scholar]
- 23.Dubik D, Shiu RP. Transcriptional regulation of c-myc oncogene expression by estrogen in hormone-responsive human breast cancer cells. J. Biol. Chem. 1988;263:12705–12708. [PubMed] [Google Scholar]
- 24.Robanus-Maandag EC, Bosch CA, Kristel PM, Hart AA, Faneyte IF, Nederlof PM, Peterse JL, van de Vijver MJ. Association of C-MYC amplification with progression from the in situ to the invasive stage in C-MYC-amplified breast carcinomas. J. Pathol. 2003;201:75–82. doi: 10.1002/path.1385. [DOI] [PubMed] [Google Scholar]
- 25.Corzo C, Corominas JM, Tusquets I, Salido M, Bellet M, Fabregat X, Serrano S, Sole F. The MYC oncogene in breast cancer progression: from benign epithelium to invasive carcinoma. Cancer Genet. Cytogenet. 2006;165:151–156. doi: 10.1016/j.cancergencyto.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Blancato J, Singh B, Liu A, Liao DJ, Dickson RB. Correlation of amplification and overexpression of the c-myc oncogene in high-grade breast cancer: FISH, in situ hybridisation and immunohistochemical analyses. Br. J. Cancer. 2004;90:1612–1619. doi: 10.1038/sj.bjc.6601703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cady NC, Strickland AD, Batt CA. Optimized linkage and quenching strategies for quantum dot molecular beacons. Mol. Cell. Probes. 2007;21:116–124. doi: 10.1016/j.mcp.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Berthois Y, Katzenellenbogen JA, Katzenellenbogen BS. Phenol red in tissue culture media is a weak estrogen: implications concerning the study of estrogen-responsive cells in culture. Proc. Natl Acad. Sci. USA. 1986;83:2496–2500. doi: 10.1073/pnas.83.8.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu I, Blackwell K, Chen S, Slingerland J. The dual ErbB1/ErbB2 inhibitor, lapatinib (GW572016), cooperates with tamoxifen to inhibit both cell proliferation- and estrogen-dependent gene expression in antiestrogen-resistant breast cancer. Cancer Res. 2005;65:18–25. [PubMed] [Google Scholar]
- 30.Wolf SF, Haines L, Fisch J, Kremsky JN, Dougherty JP, Jacobs K. Rapid hybridization kinetics of DNA attached to submicron latex particles. Nucleic Acids Res. 1987;15:2911–2926. doi: 10.1093/nar/15.7.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henry MR, Stevens PW, Sun J, Kelso DM. Real-time measurements of DNA hybridization on microparticles with fluorescence resonance energy transfer. Anal. Biochem. 1999;276:204–214. doi: 10.1006/abio.1999.4344. [DOI] [PubMed] [Google Scholar]
- 32.Brown DA, Kang SH, Gryaznov SM, DeDionisio L, Heidenreich O, Sullivan S, Xu X, Nerenberg MI. Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J. Biol. Chem. 1994;269:26801–26805. [PubMed] [Google Scholar]
- 33.Fisher TL, Terhorst T, Cao X, Wagner RW. Intracellular disposition and metabolism of fluorescently-labeled unmodified and modified oligonucleotides microinjected into mammalian cells. Nucleic Acids Res. 1993;21:3857–3865. doi: 10.1093/nar/21.16.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah YM, Al-Dhaheri M, Dong Y, Ip C, Jones FE, Rowan BG. Selenium disrupts estrogen receptor (alpha) signaling and potentiates tamoxifen antagonism in endometrial cancer cells and tamoxifen-resistant breast cancer cells. Mol. Cancer Ther. 2005;4:1239–1249. doi: 10.1158/1535-7163.MCT-05-0046. [DOI] [PubMed] [Google Scholar]
- 35.Doisneau-Sixou SF, Cestac P, Faye JC, Favre G, Sutherland RL. Additive effects of tamoxifen and the farnesyl transferase inhibitor FTI-277 on inhibition of MCF-7 breast cancer cell-cycle progression. Int. J. Cancer. 2003;106:789–798. doi: 10.1002/ijc.11263. [DOI] [PubMed] [Google Scholar]
- 36.Johnson DR, Lee PK, Holmes VF, Alvarez-Cohen L. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl. Environ. Microbiol. 2005;71:3866–3871. doi: 10.1128/AEM.71.7.3866-3871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitsuhashi M, Tomozawa S, Endo K, Shinagawa A. Quantification of mRNA in whole blood by assessing recovery of RNA and efficiency of cDNA synthesis. Clin. Chem. 2006;52:634–642. doi: 10.1373/clinchem.2005.048983. [DOI] [PubMed] [Google Scholar]
- 38.Tian X, Aruva MR, Qin W, Zhu W, Sauter ER, Thakur ML, Wickstrom E. Noninvasive molecular imaging of MYC mRNA expression in human breast cancer xenografts with a [99mTc]peptide-peptide nucleic acid-peptide chimera. Bioconjug. Chem. 2005;16:70–79. doi: 10.1021/bc0497923. [DOI] [PubMed] [Google Scholar]
- 39.Tian X, Chakrabarti A, Amirkhanov NV, Aruva MR, Zhang K, Mathew B, Cardi C, Qin W, Sauter ER, et al. External imaging of CCND1, MYC, and KRAS oncogene mRNAs with tumor-targeted radionuclide-PNA-peptide chimeras. Ann. N.Y. Acad. Sci. 2005;1059:106–144. doi: 10.1196/annals.1339.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.