Abstract
Fluorescent labeling of nucleic acids is widely used in basic research and medical applications. We describe the efficient site-specific incorporation of a fluorescent base analog, 2-amino-6-(2-thienyl)purine (s), into RNA by transcription mediated by an unnatural base pair between s and pyrrole-2-carbaldehyde (Pa). The ribonucleoside 5′-triphosphate of s was site-specifically incorporated into RNA, by T7 RNA polymerase, opposite Pa in DNA templates. The fluorescent intensity of s in RNA molecules changes according to the structural environment. The site-specific s labeling of RNA hairpins and tRNA molecules provided characteristic fluorescent profiles, depending on the labeling sites, temperature and Mg2+ concentration. The Pa-containing DNA templates can be amplified by PCR using 7-(2-thienyl)imidazo[4,5-b]pyridine (Ds), another pairing partner of Pa. This site-specific fluorescent probing by the unnatural pair system including the s-Pa and Ds-Pa pairs provides a powerful tool for studying the dynamics of the local structural features of 3D RNA molecules and their intra- and intermolecular interactions.
INTRODUCTION
Studies of the structural dynamics of biopolymers, such as nucleic acids and proteins, and their complexes in solution are keys to understanding their functions and mechanisms in living organisms. Nucleic acids, especially RNA, are extremely versatile and flexible molecules that are capable of interacting with many other molecules, and even subtle local variations in structure and dynamics are functionally significant. For detecting the local structural changes, fluorescent probes using base analogs, such as 2-aminopurine (AP) (1–6), pteridines (7–9), cyclic cytosines (10,11), 5-(fur-2-yl)uridine (12) and hydrocarbons (13,14), provide useful information that may not be obtained by a static structural analysis. The fluorescent intensity of AP at defined positions in nucleic acids varies depending on its structural environment, thus making it useful as a probe of local structural changes. For example, stacking interactions with the neighboring bases quench the AP fluorescence. Using this technique, several functional RNA molecules, such as ribozymes and aptamers, were analyzed (15–22). However, the use of fluorescent base analogs is still restricted, because their site-specific incorporation into nucleic acids relies solely on chemical synthesis. Although the combination of chemical synthesis and enzymatic ligation is now a routine and reliable procedure for DNA preparation, it is still difficult and laborious to prepare RNA fragments with long chains. Thus, the further development of the site-specific incorporation of fluorescent base analogs into RNA could expand the fluorescence techniques, which promise to be broadly applicable.
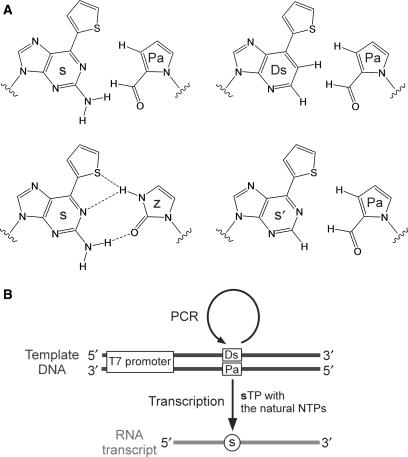
One of the attractive methods for the site-specific fluorescent labeling of RNA is the expansion of the genetic alphabet by an unnatural base-pair system (23–26). This system enables the enzymatic incorporation of extra components into RNA at desired positions by transcription mediated by the extra base pairs. Recently, we developed unnatural base pairs, such as 7-(2-thienyl)imidazo[4,5-b]pyridine (denoted by Ds) and pyrrole-2-carbaldehyde (denoted by Pa) (27), 2-amino-6-(2-thienyl)purine (denoted by s) and 2-oxopyridine (denoted by y) (28–30), and s and imidazolin-2-one (denoted by z) (31). Each base pair has a specific and characteristic selectivity in replication and transcription. For example, the Ds–Pa pair functions complementarily in replication and transcription, enabling the site-specific incorporations of Ds and Pa into DNA and RNA by polymerases. In addition, the s–y pair can be used unidirectionally in transcription for incorporating y and modified y, such as fluorophore-linked y bases, into RNA opposite s in DNA templates.
The s base is strongly fluorescent (excitation: 299 and 352 nm; emission: 434 nm; quantum yield: 41%) (32). Both the excitation and emission centers of s are shifted to longer wavelengths, relative to those of AP. In addition, the quenching of the s fluorescence in nucleic acids is sensitive to its stacking environment, but is less than that of AP, and the s fluorescence is efficiently detectable even in nucleic acids. Thus, the s base would be more useful as a fluorescent probe for analyzing the local structural dynamics of nucleic acids. For the site-specific incorporation of s into RNA, the s–z pair (Figure 1A) can be used; the substrate of s is selectively incorporated into RNA opposite z in DNA templates by T7 transcription (31). However, the transcription mediated by the s–z pair is less efficient relative to that by the natural base pairs. Furthermore, DNA templates containing the s–z pair cannot be amplified by PCR, due to its insufficient selectivity for precise replication. Therefore, the further development of unnatural base-pair systems for s incorporation into RNA should provide a powerful tool for the site-specific fluorescent labeling of RNA molecules.
Figure 1.
The unnatural base-pair system for specific replication and transcription. (A) Structures of the unnatural s–Pa, Ds–Pa, s–z and s′–Pa pairs. (B) The unnatural base-pair system for site-specific incorporation of the fluorescent s base into RNA by transcription using the Pa-containing DNA templates, which can be amplified by PCR mediated by the Ds–Pa pair.
Here, we report an unnatural base-pair system combining a novel s–Pa pair with the Ds–Pa pair (Figure 1A). We initially developed Pa as a pairing partner of Ds, and then serendipitously found that Pa in DNA fragments also functions as a template base for the site-specific incorporation of s, as well as Ds, in transcription. The s–Pa pair enables the efficient, site-specific incorporation of the fluorescent s substrate into RNA, by T7 transcription, opposite Pa in templates. Furthermore, the Pa-containing DNA templates can be amplified by PCR via the Ds–Pa pair (27) (Figure 1B). Using this system, we employed the site-specific incorporation of the fluorescent s probe to analyze the local structures of RNA hairpins with GNRA loops and tRNA molecules.
MATERIALS AND METHODS
General
Reagents and solvents were purchased from standard suppliers without further purification. Electrospray ionization mass spectra (ESI-MS) were recorded on a Waters ZMD 4000 LC/MS system. The DNA templates were chemically synthesized with an automated DNA synthesizer (model 392, PerkinElmer Applied Biosystems, Foster City, CA, USA) using the phosphoramidites of Pa (27) and the natural bases. The oligonucleotides were purified by gel electrophoresis. The substrates of s and 6-(2-thienyl)purine (s′) were chemically synthesized from its ribonucleoside (see Supplementary Data).
DNA templates for T7 transcription
Chemically synthesized DNA templates (10 μM of a 35-mer template strand and a 21-mer non-template strand for 17-mer RNA synthesis; 10 μM of a 37-mer template strand and a 23-mer non-template strand for 19-mer hairpin RNA synthesis; 5 μM of 94-mer template and non-template DNAs for tRNA synthesis) were annealed in a buffer containing 10 mM Tris-HCl (pH 7.6) and 10 mM NaCl, by heating at 95°C and slow cooling to 4°C.
T7 transcription (17-mer RNA)
Transcription was performed in a reaction buffer (20 μl) containing 40 mM Tris-HCl (pH 8.0), 24 mM MgCl2, 2 mM spermidine, 5 mM DTT and 0.01% Triton X-100 in the presence of 1 mM natural NTPs, 0 or 1 mM sTP, 2 μCi [γ-32P]GTP, 2 μM DNA template and 50 U of T7 RNA polymerase (Takara, Kyoto). By the use of [γ-32P]GTP, the transcripts were labeled only at the 5′-end, which facilitated the analyses of the yields. After an incubation at 37°C for 3 h, the reaction was quenched by the addition of a dye solution (20 μl) containing 10 M urea and 0.05% BPB. The mixture was heated at 75°C for 3 min, and the products were analyzed on a 20% polyacrylamide–7 M urea gel.
Nucleotide-composition analysis in T7 transcription (17-mer RNA)
Transcription was performed in the reaction buffer (20 μl) with 10 mM GMP, 1 mM natural NTPs, 0, 1 or 3 mM sTP, 2 μCi [α-32P]UTP, [α-32P]ATP or [α-32P]GTP (GE Healthcare), 2 μM template, and 50 U of T7 RNA polymerase (Takara) (27,28). After an incubation for 3 h at 37°C, the transcription was quenched by the addition of the dye solution. This mixture was heated at 75°C for 3 min, and then was loaded onto a 15% polyacrylamide–7 M urea gel. The full-length products were eluted from the gel with water, and were precipitated with ethanol and 0.05 A260 units of Escherichia coli tRNA. The transcripts were digested by 0.075 U/μl RNase T2 at 37°C for 2 h, in 15 mM sodium acetate buffer (pH 4.5). The digestion products were analyzed by 2D-TLC, using a Merck HPTLC plate (100 × 100 mM) (Merck, Darmstadt, Germany) with the following developing solvents: isobutyric acid/NH4OH/H2O (66:1:33 v/v/v) for the first dimension, and isopropyl alcohol/HCl/H2O (70:15:15 v/v/v) for the second dimension. The products on the gels and the TLC plates were analyzed with a Bio-imaging analyzer, BAS2500 (Fuji Film). The quantification of each spot was averaged from 3 to 9 data sets.
Preparation of the 10s and 11s RNA hairpins
The sequences of the templates for the RNA hairpins containing s are listed in the Supplementary Data. Transcription was performed in the reaction buffer with 2 mM natural NTPs, 2 mM sTP, 2 μM template and 2.5 U/μl T7 RNA polymerase. After an incubation for 3 h at 37°C, the full-length products were purified on a 15% polyacrylamide–7 M urea gel. The products were resuspended in a buffer containing 10 mM sodium phosphate (pH 7.0), 100 mM NaCl and 0.1 mM EDTA, and the amount of each RNA was determined by the absorbance at 260 nm. The solutions were diluted to 2.5 μM (∼0.5 OD260/ml) with the buffer for fluorescent and UV melting measurements.
Preparation of the 16s, 17s, 36s, 47s, 57s and 59s tRNAs
The sequences of the templates for the yeast tRNAPhe molecules containing s are listed in the Supplementary Data. In the sequences, the C2–G71 pair was replaced by G2–C71. The last two nucleosides (G and T) at the 5′-termini of the template strands were replaced with their 2′-O-methylribonucleosides, to reduce the addition of one or more non-templated nucleotides at the 3′-terminus of the nascent transcript (33). Transcription was performed in the reaction buffer with 10 mM GMP, 1 mM natural NTPs, 1 mM sTP, 0.5 μM template and 2.5 U/μl T7 RNA polymerase. After an incubation for 6 h at 37°C, the transcription was quenched by the addition of an equivalent volume of water and 1.75 volumes of the dye solution. This mixture was heated at 75°C for 3 min, and then was loaded onto a 10% polyacrylamide–7 M urea gel. The full-length products were eluted from the gel with water, and were precipitated with ethanol. The products were resuspended in 450 μl of 10 mM EDTA (pH 8) and were incubated at 75°C for 5 min. Then, using a Microcon YM-10 filter (Amicon), the buffer solution was exchanged to Tm buffer containing 50 mM sodium cacodylate (pH 7.2) and 50 mM KCl. The amounts of tRNA were determined by the absorbance at 260 nm, and the solutions were diluted to 1 μM tRNA in Tm buffers containing either 0.1 mM EDTA, 2 mM MgCl2 or 5 mM MgCl2, for fluorescent and UV melting measurements.
Fluorescent and UV melting curves
Fluorescent and UV melting profiles of each RNA hairpin or tRNA containing s at a specific position were recorded at a heating rate of 0.5°C/min from 20 to 90°C, using an FP-6500 spectrofluorimeter (JASCO) equipped with a thermoelectric cell holder, and a UV-2450 spectrophotometer (SHIMADZU), respectively. For the emission spectra, the excitation wavelength was 352 nm with a 3-nm spectral bandwidth. For baseline correction, we independently determined the temperature dependence of the fluorescence for the s ribonucleoside. Each melting temperature was calculated by using the IGOR Pro software (WaveMetrics, Inc.).
RESULTS
T7 transcription using the s–Pa pair
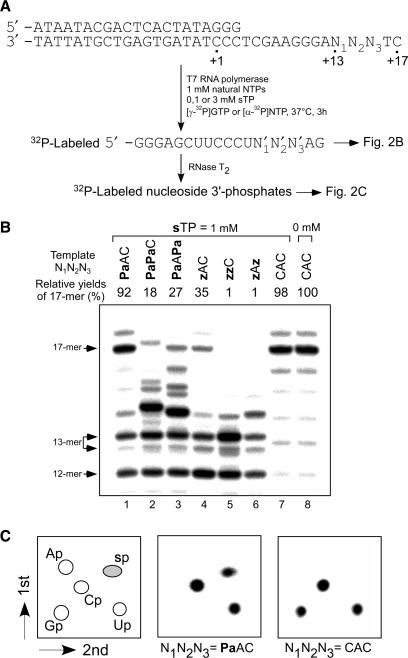
First, we examined the incorporation efficiency and selectivity of the substrate of s (sTP) using short DNA templates (35-mer) containing one or two Pa bases, in which the unnatural bases were located at complementary sites corresponding to positions 13–15 in the 17-mer transcripts (Figure 2A). The ability of Pa to function as the template base for s incorporation was compared with that of another unnatural base, z (31). After 3 h of transcription with [γ-32P] GTP, the 5′-labeled transcripts were analyzed on a gel (Figure 2B). The mobility of each of the full-length transcripts and truncated products slightly differed on the gel, depending on its base composition and sequence. In the efficient transcription (Figure 2B, lanes 1, 7 and 8), 18-mer products, which were obtained by the addition of one non-templated nucleotide to the full-length transcripts (17-mer), were observed, as in the common T7 transcription. The truncated products were also observed in transcription involving the unnatural base pairs (Figure 2B, lanes 1–6). The 13-mer truncated products resulted from pausing after the incorporation of sTP (the upper bands of the 13-mer) or the misincorporation of the natural NTPs, mainly ATP (27) (the lower bands of the 13-mer).
Figure 2.
T7 transcription mediated by the s–Pa pairing. (A) Schemes of the experiments. (B) Gel electrophoresis of transcripts generated from templates containing one or two Pa or z bases with the natural NTPs (1 mM) and sTP (1 mM). Transcripts were labeled at their 5′-termini with [γ-32P]GTP. The relative yields of each transcript were determined by comparison to the yields of native transcripts from templates consisting of the natural bases, and each yield was averaged from 3 to 4 data sets. (C) 2D-TLC analysis of the labeled ribonucleoside 3′-phosphates obtained from the nuclease digestion of the transcripts (17-mer). The transcripts were internally labeled with [α-32P]UTP. The spots on the TLC were obtained from the 17-mer fragment transcribed from the template (N1N2N3 = PaAC or CAC) in the presence of 1 mM sTP.
Despite the production of the truncated transcripts, the relative yield (92%) of the full-length transcripts containing one s base from the Pa template (Figure 2B, lane 1) was much higher than that from the z template (35%) (Figure 2B, lane 4), and as high as that obtained from the natural template with the natural NTPs (Figure 2B, lane 8). Even in transcription using templates containing two Pa bases, the full-length products were observed (Figure 2B, lanes 2 and 3), although the relative yields were lower than that of the native transcription. In contrast, transcription reactions using the templates containing two z bases did not yield full-length transcripts (Figure 2B, lanes 5 and 6). Thus, the transcription efficiency of the s–Pa pair was significantly improved, as compared to that of the s–z pair.
To assess the selectivity of the s–Pa pairing in transcription, we analyzed the nucleotide composition of the full-length transcripts obtained from the Pa, z and natural-base templates. For the analysis, the transcripts were internally labeled with either [α-32P] UTP, ATP or GTP, chosen depending on the template sequence to label the 3′-side of the incorporated s nucleoside, and then the transcripts were fully digested to nucleoside 3′-phosphates with RNase T2. The labeled nucleoside 3′-phosphates were analyzed by 2D-TLC (Figure 2C), and the nucleotide composition was quantified (Table 1).
Table 1.
Nucleotide-composition analysis of T7 transcripts
| Entry | Template N1N2N3 | [α-32P] NTP | sTP or s′TP (mM) | Composition of nucleotides incorporated as 5′ neighbor of U or A or Ga | ||||
|---|---|---|---|---|---|---|---|---|
| Ap | Gp | Cp | Up | s′p | ||||
| 1 | PaAC | UTP | sTP (1) | 0.01b [0]c (0.01)d | n.d.e [0] (<0.01) | 1.99 [2] (0.05) | 1.02 [1] (0.03) | 0.97 [1] (0.03) |
| 2 | zAC | UTP | sTP (1) | 0.01 [0] (<0.01) | 0.01 [0] (0.01) | 1.99 [2] (0.02) | 1.02 [1] (0.05) | 0.97 [1] (0.05) |
| 3 | CAC | UTP | – (0) | 0.01 [0] (<0.01) | 0.97 [1] (0.03) | 2.02 [2] (0.02) | 1.01 [1] (0.03) | n.d. [0] (–) |
| 4 | CAC | UTP | sTP (1) | 0.01 [0] (<0.01) | 0.98 [1] (0.02) | 2.03 [2] (0.03) | 0.99 [1] (0.04) | n.d. [0] (–) |
| 5 | TAC | UTP | – (0) | 1.00 [1] (0.03) | 0.01 [0] (<0.01) | 2.02 [2] (0.01) | 0.97 [1] (0.04) | n.d. [0] (–) |
| 6 | TAC | UTP | sTP (1) | 1.00 [1] (0.03) | 0.01 [0] (<0.01) | 2.01 [2] (0.01) | 0.97 [1] (0.03) | 0.01 [0] (<0.01) |
| 7 | PaAPa | UTP | sTP (1) | 0.01 [0] (0.01) | 0.01 [0] (<0.01) | 2.03 [2] (0.03) | 1.00 [1] (0.02) | 0.95 [1] (0.03) |
| 8 | PaAPa | UTP | sTP (3) | 0.01 [0] (0.01) | 0.01 [0] (0.01) | 2.01 [2] (0.01) | 0.99 [1] (0.01) | 0.98 [1] (0.01) |
| 9 | PaAPa | ATP | sTP (1) | 0.05 [0] (<0.01) | 1.06 [1] (0.07) | n.d. [0] (–) | 0.01 [0] (<0.01) | 0.87 [1] (0.07) |
| 10 | PaAPa | ATP | sTP (3) | 0.04 [0] (0.01) | 1.02 [1] (0.06) | n.d. [0] (–) | 0.01 [0] (<0.01) | 0.93 [1] (0.07) |
| 11 | CAC | ATP | sTP (1) | 0.03 [0] (<0.01) | 1.95 [2] (0.01) | n.d. [0] (–) | 0.02 [0] (0.01) | n.d. [0] (–) |
| 12 | CAC | ATP | sTP (3) | 0.03 [0] (<0.01) | 1.94 [2] (0.01) | n.d. [0] (–) | 0.02 [0] (0.01) | n.d. [0] (–) |
| 13 | PaPaC | GTP | sTP (1) | 1.99 [2] (0.05) | 1.15 [1] (0.05) | 0.02 [0] (0.02) | n.d. [0] (–) | 0.84 [1] (0.03) |
| 14 | PaPaC | GTP | sTP (3) | 1.94 [2] (0.03) | 1.13 [1] (0.07) | 0.02 [0] (0.01) | 0.01 [0] (0.02) | 0.89 [1] (0.10) |
| 15 | CAC | GTP | sTP (1) | 1.98 [2] (0.06) | 1.01 [1] (0.03) | 0.01 [1] (0.01) | 1.98 [2] (0.08) | 0.01 [1] (0.01) |
| 16 | CAC | GTP | sTP (3) | 1.99 [2] (0.04) | 0.99 [1] (0.03) | 0.01 [0] (0.01) | 1.99 [2] (0.08) | 0.02 [1] (0.01) |
| 17 | PaAC | UTP | s′TP (1) | 0.02 [0] (<0.01) | 0.01 [0] (0.01) | 2.01 [2] (0.02) | 1.01 [1] (0.01) | 0.95 [1] (0.01) |
| 18 | zAC | UTP | s′TP (1) | 0.04 [0] (<0.01) | 0.08 [0] (0.01) | 2.01 [2] (0.01) | 1.01 [1] (0.01) | 0.85 [1] (0.01) |
| 19 | CAC | UTP | s′TP (1) | 0.01 [0] (<0.01) | 0.98 [1] (0.01) | 2.01 [2] (0.02) | 1.00 [1] (0.02) | n.d. [0] (–) |
aComposition of nucleotides incorporated as 5′ neighbor of U (Entries 1–8 and 17–19), A (Entries 9–12) or G (Entries 13–16), as shown in Figure 2.
bThe values were determined using the following formula: (radioactivity of each nucleotide)/[total radioactivity of all nucleotides (3′-monophosphates)] × (total number of nucleotides at 5′ neighbor of [α-32P]NTP).
cThe theoretical number of each nucleotide is shown in brackets.
dSDs are shown in parentheses.
eNot detected.
The results confirmed the faithful s incorporation using the template containing one Pa base. The labeled s-nucleoside 3′-phosphate was observed on the 2D-TLC only when the template containing Pa was used (Figure 2C, N1N2N3 = PaAC). The selectivity (97%) of the s incorporation opposite Pa (Table 1, entry 1) was as high as that mediated by the s–z pair (Table 1, entry 2) and that of the natural transcription, and no misincorporation of sTP opposite natural bases was observed (Figure 2C, N1N2N3 = CAC and Table 1, entries 4, 6). In transcription using the templates containing two Pa bases (N1N2N3 = PaAPa and PaPaC), the selectivity of the s incorporation at the second positions decreased to 84–87% (Table 1, entries 9, 13), although the selectivity of the s incorporation at the first position was high (95%) (Table 1, entry 7). This selectivity was improved, to some extent (89–93%), by increasing the concentration (3 mM) of sTP (Table 1, entries 10, 14).
To understand the role of the 2-amino group of s in the pairing with Pa in transcription, we also chemically synthesized the substrate of 6-(thienyl)purine (s′) (34) (Figure 1), which lacks the 2-amino group of s, and examined the efficiency and selectivity of the s′-incorporation into RNA opposite Pa or z. The s′ substrate was also efficiently incorporated into RNA opposite Pa by transcription (data not shown). The selectivity of the s′ incorporation opposite Pa (95%) (Table 1, entry 17) was as high as that of the s incorporation opposite Pa. However, the selectivity of the s′–z pairing was decreased by 85%, suggesting the presence of a hydrogen-bonding interaction between the 2-amino group of s and the 2-keto of z. In contrast, we have no evidence for a possible interaction between the 2-amino group of s and the 2-aldehyde group of Pa in transcription. The DNA fragment containing the s–Pa pair exhibited thermal stability similar to that of the DNA fragment with the s′–Pa pair (data not shown).
Site-specific fluorescent s labeling of a GNRA hairpin
Using the s–Pa pair, we examined the site-specific incorporation of s into RNA hairpins by T7 transcription to demonstrate the potential of the fluorescent s base as a probe. We incorporated s into the loop region of RNA hairpins containing GNRA loops (where N can be any nucleotide and R is either G or A). RNA hairpins with GNRA loops, such as GAAA, GAGA and GCAA loops, exhibit high thermal stability and are extremely common in biologically active RNA molecules. NMR studies (35,36) and fluorescent probing (16,20) of the hairpins indicated that the GNRA loops contain a sheared G–A pair between the first G and fourth A in the loop. Although the third base (R) is always stacked with the fourth A in the loop, the second base (N) is less ordered than the third base. Thus, it is commonly believed that the second base is apt to be exposed on the outside of the loop, where it would be accessible for interactions with other molecules.
We prepared hairpin fragments containing s at the second or third position in the GNRA loop (Figure 3A and B) by T7 transcription using Pa templates with sTP and the natural NTPs. Temperature-dependent melting profiles of each transcript (2.5 μM) were measured, using the fluorescence emission at 434 nm of s (excited at 352 nm) and the UV absorbance at 260 nm (Figure 3C). Since the quenching of the s fluorescence by collision events with the solvent increases at higher temperatures, the fluorescence-monitored melting curves of each RNA transcript were normalized by that of the nucleoside monomer of s (see Supplementary Data). The s incorporation into the loop only slightly reduced the thermal stability of the hairpins, by 1.4–1.5°C, as compared to the unmodified hairpin containing the GAAA loop (melting temperature (Tm) = 68.2°C). Thus, the substitution of the second or third base in the loop with s does not induce any substantial change in the entire hairpin structure.
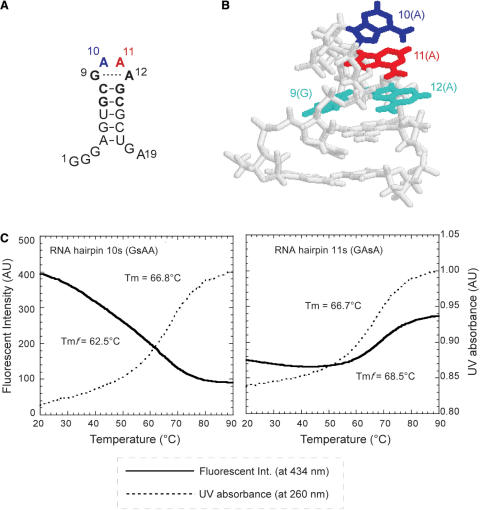
Figure 3.
Incorporation of the fluorescent s base into GNRA hairpins. (A) The secondary structure of the RNA hairpin with a GAAA loop. The second A and third A in the loop are shown in blue and red, respectively. (B) The 3D structure of the GAAA-loop hairpin (35). (C) The profiles of the fluorescent intensity at 434 nm (solid lines) and the UV absorption at 260 nm (dotted lines) of the RNA hairpin with a GsAA (RNA hairpin 10s) or GAsA (RNA hairpin 11s) loop. Tm and Tmf values were obtained from the UV melting and fluorescent intensity profiles, respectively.
The GsAA and GAsA hairpins (10s and 11s) showed characteristic fluorescent profiles (Figure 3C), which reflect the general GNRA loop structures. At physiological temperatures, the fluorescent intensity of the GsAA loop (10s) was much larger than that of the GAsA loop (11s), even though the s base was next to the first G base in the GsAA loop, and in general, G quenches fluorescence most efficiently among the natural bases. The strong fluorescence emitted by the GsAA loop gradually decreased when the temperature was increased. On the contrary, the intensity of the GAsA loop quickly increased at around 60–76°C. These observations indicate that the second s base in the GsAA loop is exposed to the solvent at physiological temperatures, and then the non-specific interactions of s with the neighboring bases increase gradually by the denaturation of the hairpin structure with an increase in temperature. In contrast, the third s base in the GAsA loop stacks with the fourth A base at physiological temperatures, and this base stacking unfolds upon the denaturation of the hairpin structure by increasing the temperature.
The melting temperatures obtained from the fluorescent profiles (denoted by Tmf) also provide valuable information about the thermal stability of each local region. The Tmf value (62.5°C) of the GsAA loop was much lower than the UV melting temperature (66.8°C) of the hairpin. This shows the high flexibility of the second base in the GsAA loop. In contrast, the Tmf value (68.5°C) of the GAsA loop was higher than the UV melting temperature (66.7°C), indicating that the stacking of the third base in the GAsA loop strongly contributes to the stability of the entire hairpin structure. These results highlight the potential of the s base as a fluorescent probe.
Site-specific fluorescent s labeling of tRNA
Next, the ability of the fluorescent s probe was tested in a more intricate RNA molecule, tRNA. We incorporated one s base into yeast tRNAPhe at six specific positions by T7 transcription, and examined the Mg2+-induced and temperature-dependent tRNA folding by analyzing the fluorescent properties of s at each site in the tRNA. For the s incorporation into the tRNA, we chose six sites: positions 16 and 17 in the D-loop (tRNA 16s and 17s), 36 in the anticodon (tRNA 36s), 47 in the extra-loop (tRNA 47s) and 57 and 59 in the TΨC-loop (tRNA 57s and 59s), where each original base does not pair with any other base (Figure 4).
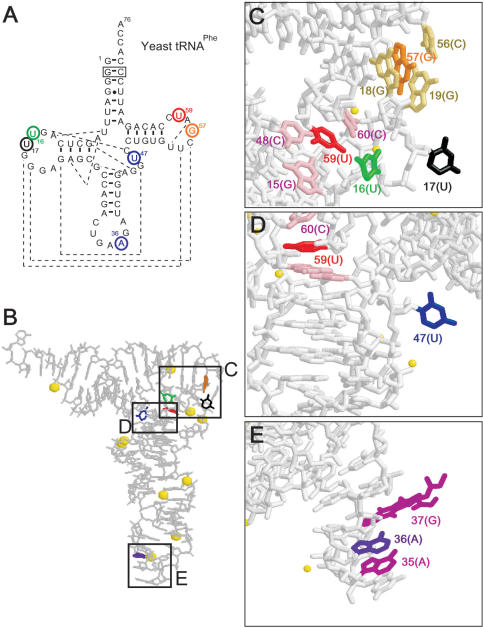
Figure 4.
The s incorporation sites in yeast tRNAPhe and the structure of the tRNA. (A) The secondary structure of the original tRNA transcript. The positions substituted with s are circled. The broken lines show base–base interactions for the 3D structure (37). The boxed G–C pair was changed from the original C–G pair, but this mutation does not significantly alter the original tRNA structure. (B–E) The deep-colored bases were substituted with s, which stacks with the light-colored bases, and the yellow spheres represent Mg2+.
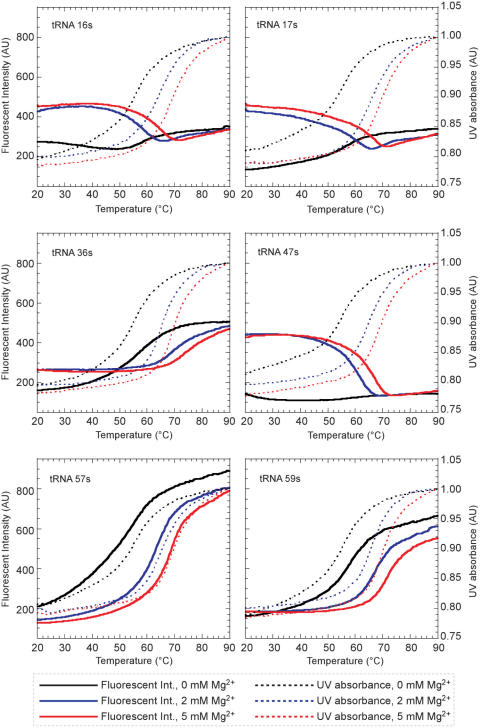
Temperature-dependent melting profiles of the fluorescence emission at 434 nm of s (excited at 352 nm) and the UV absorbance at 260 nm of each tRNA transcript (1 μM) were measured (Figure 5 and Table 2). The UV melting temperature of each tRNA transcript containing s (64.6–66.4°C in 2 mM MgCl2) was as high as that of the natural tRNA transcript (65.5°C in 2 mM MgCl2), suggesting that the substitution of s at these positions did not significantly destabilize the global tRNA structure.
Figure 5.
The fluorescent intensity and UV absorption profiles of tRNA molecules containing s at specific positions. Melting curves obtained by the changes in the fluorescent intensities at 434 nm (solid lines) and in the UV absorbance at 260 nm (dotted lines) of the tRNAs containing s at various positions.
Table 2.
Melting temperatures obtained from fluorescent and UV profiles of yeast tRNAPhe containing s at specific sites
| Tmf values obtained from fluorescent profiles (°C) | Tm values obtained from UV profiles (°C) | |||||
|---|---|---|---|---|---|---|
| 0 mM Mg2+ | 2 mM Mg2+ | 5 mM Mg2+ | 0 mM Mg2+ | 2 mM Mg2+ | 5 mM Mg2+ | |
| Original tRNA | – | – | – | 56.1 | 65.5 | 70.1 |
| tRNA 16s | n.c. | 58.0 | 64.0 | 55.6 | 64.6 | 69.1 |
| tRNA 17s | n.c. | 59.6 | 65.5 | 55.7 | 65.6 | 69.1 |
| tRNA 36s | 57.0 | 69.5 | 74.0 | 55.2 | 65.2 | 69.7 |
| tRNA 47s | n.c. | 61.1 | 66.0 | 55.3 | 65.2 | 68.8 |
| tRNA 57s | 54.5 | 63.6 | 68.5 | 55.8 | 65.1 | 69.6 |
| tRNA 59s | 58.1 | 67.1 | 71.5 | 55.8 | 66.4 | 70.3 |
Fluorescent and UV profiles were obtained using each tRNA transcript (1 μM) in 50 mM sodium cacodylate (pH 7.2) and 50 mM KCl with or without MgCl2. Each melting temperature was calculated by using the IgorPro software (WaveMetrics). n.c.: not calculated.
As in the case of the simple GNRA hairpin, each tRNA containing s at the specific position also displayed characteristic fluorescent intensity changes that reflected the local structural features (Figure 5). The fluorescent profiles in the presence of Mg2+ (2 and 5 mM) clearly fall into two groups: one includes tRNA 16s, 17s and 47s (group 1), and the other includes tRNA 36s, 57s and 59s (group 2). In the presence of 2 or 5 mM Mg2+, the intensities of the s fluorescence of group 1 at low temperature were 1.7- to 3.4-fold larger than those of group 2. The fluorescent intensity of group 1 decreased when the temperature was increased, but the fluorescent intensity of group 2 increased at higher temperatures. These results indicate that the s base at position 16, 17 or 47 is exposed to the solvent at physiological temperatures, and the non-specific interactions of s with the other bases increase upon the denaturation of the folded structure with an increase in temperature. In contrast, the s base in position 36, 57 or 59 stacks with the neighboring bases in the folded structure, and the base stacking is gradually denatured by increasing the temperature. These speculations are quite consistent with the conformations of the original bases at each site in the crystal structure of the tRNA (Figure 4B–E) (37) and other structural analyses depending on Mg2+ concentrations (38,39). The fluorescence intensities of tRNA 16s, 17s and 47s drastically increased by the addition of Mg2+ at a physiological temperature, indicating that the bases at these positions are kept outside by Mg2+ binding to the L-shaped tRNA.
Furthermore, the Tmf values obtained from the fluorescent profiles reflect the stability of each local structure in the tRNA, by comparison with the Tm values obtained from the UV melting profiles. For example, the Tmf value from the fluorescent profile (69.5°C at 2 mM MgCl2) of tRNA 36s was higher than that from the UV profile (65.2°C at 2 mM MgCl2), indicating the increased stability of the anticodon stem-loop relative to the stability of the entire tRNA structure. In contrast, the low stabilities of tRNA 16s (Tmf = 58.0°C at 2 mM MgCl2) and 17s (Tmf = 59.6°C at 2 mM MgCl2) suggest that the partial structure involving the D-loop may be a fragile region within the L-shaped tRNA. Further analyses using these tRNAs will provide valuable information about the dynamics of RNA structures and the interactions with other molecules in aminoacylation and translation.
DISCUSSION
In this study, we have described the selective and effective site-specific incorporation of the fluorescent s probe into RNA by T7 transcription mediated by the s–Pa pair. We initially developed Pa as a pairing partner of another unnatural hydrophobic base, Ds, and DsTP was efficiently and selectively incorporated into RNA opposite Pa (27). We then found that Pa can also function as a template base for the site-specific incorporation of the fluorescent s base into RNA by transcription. The rational replacement of the natural bases in RNA molecules with the fluorescent s base enables pinpoint structural analysis. The fluorescent intensity of s in RNA molecules sensitively decreases with increasing stacking interactions with neighboring bases, reflecting its local structural features and the structural changes of the RNA molecules. Furthermore, the Tmf values obtained from the fluorescent profiles provide useful information about the local structural stability at the s incorporation sites in the RNA molecules. In addition to the site-specific s incorporation into RNA by T7 transcription, the DNA templates containing Pa can be amplified by PCR using the Ds–Pa pair (27). Thus, the fluorescent s probing by the unnatural base-pair system combining the s–Pa and Ds–Pa pairs provides a powerful tool for studying the dynamics of local conformational changes at a defined position within a large RNA molecule.
Our findings using this series of unnatural bases, s, s′, Ds, Pa and z, also provide a clue about the mechanisms of base pairing in transcription. In the previous report on the Ds–Pa pair (27), we showed that the non-hydrogen-bonded base-pair functions in transcription, suggesting the importance of the shape complementarity between pairing bases, as shown in replication by Kool et al. (40). Since s′ is the analog of Ds in shape, both s′ and Ds sterically fit with Pa, resulting in the high selectivity of the s′–Pa in transcription. In addition, the selectivity of the s incorporation opposite Pa was as high as that of the s′ incorporation, and thus the 2-amino group of s is not essential for the specific pairing with Pa. Even though the amino group of s may clash with the aldehyde group of Pa in the Watson–Crick base-pair geometry, T7 transcription might tolerate such a disparity between pairing bases in the selectivity. Although the hydrogen-bonding interactions may not be absolutely required for the selective base pairing in transcription, the comparison between the s–z and s′–z pairings showed that the hydrogen bonds also assist in increasing the base-pair selectivity. As for the transcription efficiency, our results indicate that the hydrophobicity of the template base is also important. Although the shape of Pa is similar to that of z, the transcription efficiency using the Pa template is much higher than that using the z template. This might be because of the higher hydrophobicity of Pa relative to z, suggesting that the hydrophobicity of the template base is favorable to the interaction with T7 RNA polymerase. In fact, the crystal structures of T7 RNA polymerase elongation complexes revealed the stacking interaction between the template bases and hydrophobic amino acid residues, such as the template (n − 1) base with Y639 and the template (n + 1) base with F644 (41,42). Additional studies using this series of unnatural bases in combination with other base analogs (23–26,43,44) could provide further understanding of the transcription mechanisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan, and by a Grant-in-Aid for Scientific Research (KAKENHI 19201046, 17750152, 18710197 and 15350097) from the Ministry of Education, Culture, Sports, Science and Technology. Funding to pay the Open Access publication charges for this article was provided by RIKEN Genomic Sciences Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ward DC, Reich E. Fluorescence studies of nucleotides and polynucleotides. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- 2.Patel N, Berglund H, Nilsson L, Rigler R, McLaughlin LW, Graslund A. Thermodynamics of interaction of a fluorescent DNA oligomer with the anti-tumour drug netropsin. Eur. J. Biochem. 1992;203:361–366. doi: 10.1111/j.1432-1033.1992.tb16558.x. [DOI] [PubMed] [Google Scholar]
- 3.Stivers JT. 2-Aminopurine fluorescence studies of base stacking interactions at abasic sites in DNA: metal-ion and base sequence effects. Nucleic Acids Res. 1998;26:3837–3844. doi: 10.1093/nar/26.16.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holtz B, Klimasauskas S, Serva S, Weinhold E. 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res. 1998;26:1076–1083. doi: 10.1093/nar/26.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rachofsky EL, Osman R, Ross JBA. Probing structure and dynamics of DNA with 2-aminopurine: effects of local environment on fluorescence. Biochemistry. 2001;40:946–956. doi: 10.1021/bi001664o. [DOI] [PubMed] [Google Scholar]
- 6.Jiao Y, Stringfellow S, Yu H. Distinguishing “looped-out” and “stacked-in” DNA bulge conformation using fluorescent 2-aminopurine replacing a purine base. J. Biomol. Struct. Dyn. 2002;19:929–934. doi: 10.1080/07391102.2002.10506795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawkins ME, Pfleiderer W, Jungmann O, Balis FM. Synthesis and fluorescence characterization of pteridine adenosine nucleoside analogs for DNA incorporation. Anal. Biochem. 2001;298:231–240. doi: 10.1006/abio.2001.5399. [DOI] [PubMed] [Google Scholar]
- 8.Stanley RJ, Hou Z, Yang A, Hawkins ME. The two-photon excitation cross section of 6MAP, a fluorescent adenine analogue. J. Phys. Chem. B. 2005;109:3690–3695. doi: 10.1021/jp0455982. [DOI] [PubMed] [Google Scholar]
- 9.Hawkins ME. Synthesis, purification and sample experiment for fluorescent pteridine-containing DNA: tools for studying DNA interactive systems. Nat. Protoc. 2007;2:1013–1021. doi: 10.1038/nprot.2007.150. [DOI] [PubMed] [Google Scholar]
- 10.Sandin P, Wilhelmsson LM, Lincoln P, Powers VEC, Brown T, Albinsson B. Fluorescent properties of DNA base analogue tC upon incorporation into DNA – negligible influence of neighboring bases on fluorescence quantum yield. Nucleic Acids Res. 2005;33:5019–5025. doi: 10.1093/nar/gki790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandin P, Lincoln P, Brown T, Wilhelmsson LM. Synthesis and oligonucleotide incorporation of fluorescent cytosine analogue tC: a promising nucleic acid probe. Nat. Protoc. 2007;2:615–623. doi: 10.1038/nprot.2007.80. [DOI] [PubMed] [Google Scholar]
- 12.Greco NJ, Tor Y. Furan decorated nucleoside analogues as fluorescent probes: synthesis, photophysical evaluation, and site-specific incorporation. Tetrahedron. 2007;63:3515–3527. doi: 10.1016/j.tet.2007.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J, Strässler C, Tahmassebi D, Kool ET. Libraries of composite polyfluors built from fluorescent deoxyribosides. J. Am. Chem. Soc. 2002;124:11590–11591. doi: 10.1021/ja027197a. [DOI] [PubMed] [Google Scholar]
- 14.Wilson JN, Kool ET. Fluorescent DNA base replacements: reporters and sensors for biological systems. Org. Biomol. Chem. 2006;4:4265–4274. doi: 10.1039/b612284c. [DOI] [PubMed] [Google Scholar]
- 15.Lacourciere KA, Stivers JT, Marino JP. Mechanism of neomycin and Rev peptide binding to the Rev responsive element of HIV-1 as determined by fluorescence and NMR spectroscopy. Biochemistry. 2000;39:5630–5641. doi: 10.1021/bi992932p. [DOI] [PubMed] [Google Scholar]
- 16.Menger M, Eckstein F, Porschke D. Dynamics of the RNA hairpin GNRA tetraloop. Biochemistry. 2000;39:4500–4507. doi: 10.1021/bi992297n. [DOI] [PubMed] [Google Scholar]
- 17.Rist MJ, Marino JP. Association of an RNA kissing complex analyzed using 2-aminopurine fluorescence. Nucleic Acids Res. 2001;29:2401–2408. doi: 10.1093/nar/29.11.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walter NG, Harris DA, Pereira MJ, Rueda D. In the fluorescent spotlight: global and local conformational changes of small catalytic RNAs. Biopolymers. 2002;61:224–242. doi: 10.1002/bip.10144. [DOI] [PubMed] [Google Scholar]
- 19.Harris DA, Rueda D, Walter NG. Local conformational changes in the catalytic core of the trans-acting hepatitis delta virus ribozyme accompany catalysis. Biochemistry. 2002;41:12051–12061. doi: 10.1021/bi026101m. [DOI] [PubMed] [Google Scholar]
- 20.Barrick JE, Roberts RW. Achieving specificity in selected and wild-type N peptide-RNA complexes: the importance of discrimination against noncognate RNA targets. Biochemistry. 2003;42:12998–13007. doi: 10.1021/bi035163p. [DOI] [PubMed] [Google Scholar]
- 21.Austin RJ, Xia TB, Ren JS, Takahashi TT, Roberts RW. Differential modes of recognition in N peptide-boxB complexes. Biochemistry. 2003;42:14957–14967. doi: 10.1021/bi0351312. [DOI] [PubMed] [Google Scholar]
- 22.Jeong S, Sefcikova J, Tinsley RA, Rueda D, Walter NG. Trans-acting hepatitis delta virus ribozyme: catalytic core and global structure are dependent on the 5′ substrate sequence. Biochemistry. 2003;42:7727–7740. doi: 10.1021/bi034627g. [DOI] [PubMed] [Google Scholar]
- 23.Benner SA, Burgstaller P, Battersby TR, Jurczyk S. Did the RNA world exploit an expanded genetic alphabet? In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 163–181. [Google Scholar]
- 24.Kool ET. Synthetically modified DNAs as substrates for polymerases. Cur. Opin. Chem. Biol. 2000;4:602–608. doi: 10.1016/s1367-5931(00)00141-1. [DOI] [PubMed] [Google Scholar]
- 25.Henry AA, Romesberg FE. Beyond A, C, G and T: augmenting nature's alphabet. Cur. Opin. Chem. Biol. 2003;7:727–733. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 26.Hirao I. Unnatural base pair systems for DNA/RNA-based biotechnology. Cur. Opin. Chem. Biol. 2006;10:622–627. doi: 10.1016/j.cbpa.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 27.Hirao I, Kimoto M, Mitsui T, Fujiwara T, Kawai R, Sato A, Harada Y, Yokoyama S. An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nat. Methods. 2006;3:729–735. doi: 10.1038/nmeth915. [DOI] [PubMed] [Google Scholar]
- 28.Hirao I, Ohtsuki T, Fujiwara T, Mitsui T, Yokogawa T, Okuni H, Nakayama K, Takio K, Yabuki T, et al. An unnatural base pair for incorporating amino acid analogs into proteins. Nat. Biotechnol. 2002;20:177–182. doi: 10.1038/nbt0202-177. [DOI] [PubMed] [Google Scholar]
- 29.Moriyama K, Kimoto M, Mitsui T, Yokoyama S, Hirao I. Site-specific biotinylation of RNA molecules by transcription using unnatural base pairs. Nucleic Acids Res. 2005;33:e129. doi: 10.1093/nar/gni128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawai R, Kimoto M, Ikeda S, Mitsui T, Endo M, Yokoyama S, Hirao I. Site-specific fluorescent labeling of RNA molecules by specific transcription using unnatural base pairs. J. Am. Chem. Soc. 2005;127:17286–17295. doi: 10.1021/ja0542946. [DOI] [PubMed] [Google Scholar]
- 31.Hirao I, Harada Y, Kimoto M, Mitsui T, Fujiwara T, Yokoyama S. A two-unnatural-base-pair system toward the expansion of the genetic code. J. Am. Chem. Soc. 2004;126:13298–13305. doi: 10.1021/ja047201d. [DOI] [PubMed] [Google Scholar]
- 32.Mitsui T, Kimoto M, Kawai R, Yokoyama S, Hirao I. Characterization of fluorescent, unnatural base pairs. Tetrahedron. 2007;63:3528–3537. [Google Scholar]
- 33.Kao C, Rüdisser S, Zheng M. A simple and efficient method to transcribe RNAs with reduced 3′ heterogeneity. Methods. 2001;23:201–205. doi: 10.1006/meth.2000.1131. [DOI] [PubMed] [Google Scholar]
- 34.Hocek M, Holý A, Votruba I, Dvořáková H. Cytostatic 6-arylpurine nucleosides III. Synthesis and structure-activity relationship study in cytostatic activity of 6-aryl-, 6-hetaryl- and 6-benzylpurine ribonucleosides. Collect. Czech. Chem. Commun. 2001;66:483–499. [Google Scholar]
- 35.Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 36.Jucker FM, Heus HA, Yip PF, Moors EH, Pardi A. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- 37.Jovine L, Djordjevic S, Rhodes D. The crystal structure of yeast phenylalanine tRNA at 2.0 Å resolution: cleavage by Mg2+ in 15-year old crystals. J. Mol. Biol. 2000;301:401–414. doi: 10.1006/jmbi.2000.3950. [DOI] [PubMed] [Google Scholar]
- 38.Shelton VM, Sosnick TR, Pan T. Altering the intermediate in the equilibrium folding of unmodified yeast tRNAPhe with monovalent and divalent cations. Biochemistry. 2001;40:3629–3638. doi: 10.1021/bi002646+. [DOI] [PubMed] [Google Scholar]
- 39.Serebrov V, Clarke RJ, Gross HJ, Kisselev L. Mg2+-induced tRNA folding. Biochemistry. 2001;40:6688–6698. doi: 10.1021/bi002241p. [DOI] [PubMed] [Google Scholar]
- 40.Morales JC, Kool ET. Efficient replication between non-hydrogen-bonded nucleoside shape analogs. Nat. Struct. Biol. 1998;5:950–954. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 41.Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, Yokoyama S. Structure of a T7 RNA polymerase elongation complex at 2.9 Å resolution. Nature. 2002;420:43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- 42.Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 43.Pochet S, D’Ari R. Synthesis and enzymatic polymerization of 5-amino-1-(2′-deoxy-β-D-ribofuranosyl)imidazole-4-carboxamide-5′-triphosphate. Nucleic Acids Res. 1990;18:7127–7131. doi: 10.1093/nar/18.23.7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Negishi K, Williams DM, Inoue Y, Moriyama K, Brown DM, Hayatsu H. The mechanism of mutation induction by a hydrogen bond ambivalent, bicyclic N4-oxy-2′-deoxycytidine in Escherichia coli. Nucleic Acids Res. 1997;25:1548–1552. doi: 10.1093/nar/25.8.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.