Abstract
Silencing of genomic repeats, including transposable elements, in Drosophila melanogaster is mediated by repeat-associated short interfering RNAs (rasiRNAs) interacting with proteins of the Piwi subfamily. rasiRNA-based silencing is thought to be mechanistically distinct from both the RNA interference and microRNA pathways. We show that the amount of rasiRNAs of a wide range of retroelements is drastically reduced in ovaries and testes of flies carrying a mutation in the spn-E gene. To address the mechanism of rasiRNA-dependent silencing of retrotransposons, we monitored their chromatin state in ovaries and somatic tissues. This revealed that the spn-E mutation causes chromatin opening of retroelements in ovaries, resulting in an increase in histone H3 K4 dimethylation and a decrease in histone H3 K9 di/trimethylation. The strongest chromatin changes have been detected for telomeric HeT-A elements that correlates with the most dramatic increase of their transcript level, compared to other mobile elements. The spn-E mutation also causes depletion of HP1 content in the chromatin of transposable elements, especially along HeT-A arrays. We also show that mutations in the genes controlling the rasiRNA pathway cause no derepression of the same retrotransposons in somatic tissues. Our results provide evidence that germinal Piwi-associated short RNAs induce chromatin modifications of their targets.
INTRODUCTION
A high level of transposable element expression is usually deleterious for the organism, leading to mutations and chromosomal rearrangements. Therefore, activity of mobile elements is thought to be under keen cellular control. Silencing of Drosophila selfish elements is realized through the short RNA species, called repeat associated short interfering RNAs (rasiRNAs) (1–5) and also Piwi-interacting RNAs (piRNAs) (6). piRNAs play evolutionarily conserved roles in the regulation of transposable elements in insects, mammals and zebrafish (7–9) and are accumulated specifically in the germline (9–12).
In Drosophila, the rasiRNA pathway requires members of the ‘Piwi subfamily’ of Argonaute proteins Piwi, Aubergine (Aub) and Ago-3 (2–6) but not the ‘Argonaute subfamily’ members, Ago1 or Ago2 (3), which guide microRNA and siRNA functions, respectively (13). rasiRNAs of 24–28 nt in length are longer than 21–22 nt siRNAs derived from dsRNA or 21–23 nt endogenous microRNAs (1,2). The increased length of rasiRNAs has aroused a suggestion of a peculiar mechanism of their formation (2). In flies neither Dicer-1, which makes microRNAs, nor Dicer-2, which produces siRNAs, are implicated in rasiRNA formation (3). Recent publications support a model in which discrete heterochromatic loci produce rasiRNAs that are predominantly antisense to transposons. The antisense rasiRNAs are bound by Piwi and Aub proteins and guide formation of sense rasiRNAs by cleavage of sense transposon transcripts (5,6).
It was demonstrated that short interfering RNAs are implicated in chromatin modifications, such as methylation of histone H3 K9, in yeast, plants and animal somatic cells (14–17). However, it remains unknown whether chromatin-based silencing of selfish elements may be realized in the germline by Piwi-interacting RNAs, in particular by rasiRNAs in flies.
We show that the significantly reduced abundance of rasiRNAs derived from a wide range of transposable elements in spn-E mutant ovaries is accompanied by the increase of H3 K4 dimethylation, decrease of H3 K9 di/trimethylation and depletion of HP1 content in the chromatin of retrotransposons. We demonstrate that rasiRNA-mediated silencing of tested retrotransposons takes place in ovaries, where it is necessary to protect the genome against transposon-induced mutations in progeny, but not in somatic tissues.
MATERIALS AND METHODS
Drosophila strains
Strains bearing spn-E1, piwi2 and armi1 mutations were ru1 st1 spn-E1 e1 ca1/TM3, Sb1 es (point mutation in helicase domain of Spn-E), ru1 st1 spn-Ehls3987 e1 ca1/TM3, Sb1 es (P-element insertion into spn-E) (18,19), piwi2/CyO (P-ry11 transposon insertion) (20) and armi1/TM3 (P-element insertion) (21), respectively. P-element transformed flies carrying the copiaLTR-lacZ construct were kindly provided by E. G. Pasyukova. Discrimination in X-gal staining experiment of homo- and heterozygous larvae carrying spn-E1, piwi2 and armi1 mutations was done using GFP-expressing balancers CyO, P{w+m = hsp70: GAL4}P{w+m = UAS: GFP} and TM3, Sb1 es P{w+m = hsp70: GAL4}P{w+m = UAS: GFP}.
RT–PCR analysis
Total RNA was isolated from dissected ovaries or carcasses using Trizol reagent (Gibco BRL). The first strand of cDNA was synthesized using SuperScript II reverse transcriptase (Gibco BRL) and oligo(dT) primer or specific primer according to the manufacturer's instructions. cDNAs were analyzed by real-time quantitative PCR using SYBR Green. For PCR the following primers were used: 5′-CCGTGGTCAACTTCACCAGCTC-3′ (adh d2) and 5′-TCCAACCAGGAGTTGAACTTGTGC-3′ (adh r2), corresponding to GenBank sequence AE003410.1 for Adh gene; 5′-TCCGCCCAGCATACAGGC-3′ (rp49 s2) and 5′-CAATCCTCGTTGGCACTCACC-3′ (rp49 as2), corresponding to GenBank sequence Y13939 for rp49 gene; 5′-GCATGAGAGGTTTGGCCATATAAGC-3′ (cop-s) and 5′-GGCCCACAGACATCTGAGTGTACTACA-3′ (cop-as), corresponding to GenBank sequence XO4456 for copia; 5′-CGCAAAGACATCTGGAGGACTACC-3′ (Het-s2) and 5′-TGCCGACCTGCTTGGTATTG-3′ (Het-as2), corresponding to GenBank sequence U06920 for HeT-A; 5′-TGAAATACGGCATACTGCCCCCA-3′ (I el s2) and 5′-GCTGATAGGGAGTCGGAGCAGATA-3′ (I el as2), corresponding to GenBank sequence M14954 for I element.
X-gal staining and β-gal activity assay
X-gal staining and β-gal activity assays were performed according to protocols described previously (2,22). Samples containing 5–15 pairs of ovaries dissected from 1 to 3-days-old females or 4–15 carcasses were used for β-gal activity assay. Measurements of β-gal activity were normalized to the total protein evaluated by the Bio-Rad protein assay kit.
Short RNA cloning and annotation
RNA preparation was performed as previously described (23). Total RNA was isolated from adult ovaries and testes. Cloning of miRNAs was performed as described (24). Characterization of cloned small RNAs was performed using local NCBI-BLAST 2.2.13 (25) against the canonical sequences of transposable elements (http://www.fruitfly.org/p_disrupt/datasets/ASHBURNER/D_mel_transposon_sequence_set.fasta); Su(Ste) repeats (GenBank accession no. X59157|H-, Z11734|H- and Z11735|H-); miRNAs (http://microrna.sanger.ac.uk/sequences/, Release 8.0), tRNA (http://lowelab.ucsc.edu/GtRNAdb/Dmela/) and rRNA (GenBank accession no. M21017). Only hits with 95% and higher similarity to transposable elements and Su(Ste) sequences and 100% similarity to other sequences were used. Parsing of results was done using corresponding BioPerl modules (26).
Chromatin IP assay
Ovaries were dissected from 1 to 10-days-old females in 1X PBS and stored in 1.5 ml tube on ice during isolation (up to 2 h). PBS solution was removed after centrifugation (3500 r.p.m. 1–2 min). 10 mg of material (about 150 ovaries or 100 carcasses) was used for one IP reaction. The chromatin IP assay was performed as described previously (27), using polyclonal rabbit antibodies (Upstate): Anti-dimethyl-Histone H3 Lys4 (#07-030), Anti-dimethyl-Histone H3 Lys9 (#07-441), Anti-trimethyl-Histone H3 Lys9 (#07-523) and anti-HP1 (PRB-291C Covance innovative). Anti-TAF1 was kindly provided by G. Cavalli. DNA precipitates were amplified by semiquantitative PCR in the presence of αP32 dATP or real-time quantitative PCR. PCR product quantities were normalized to input and relations to a fragment of intergenic spacer in the 60D region were calculated. No identified or predicted genes are located 2.5 kb upstream and 4.3 kb downstream of the 60D amplified fragment. The TRANSFAC database search found no binding sites for any known chromatin proteins and transcriptional factors in the fragment. Final enrichment values of sample PCR products were calculated using the following expression: E(product)sample *E(60D)input/E(60D)sample *E(product)input. The following primers were used for PCR analysis in ChIP: 5′-CAACACTACTTTATATTTGATATGAATGGCC-3′ and 5′-CGAAAGGGGGATGTGCTGC-3′ for amplification of the promoter region of copiaLTR-lacZ construct; 5′-CAACACTACTTTATATTTGATATGAATGGCC-3′/5′-GCGTACTTCTCGCCATCAAACG-3′ and cop-s/cop-as (see above) for endogenous copia promoter region and ORF, respectively; 5′-ACCACGCCCAACCCCCAA-3′/5′-GCTGGTGGAGGTACGGAGACAG-3′ and Het-s2/Het-as2 (see above), corresponding to HeT-A promoter region and ORF, respectively; 5′-CGTGCCTCTCAGTCTAAAGCCTC-3′/5′-CCCGGATTAGCGGTATTGTTGTT-3′ and I el s2/I el as2 (see above), corresponding to I element promoter and ORF, respectively; adh d2 and adh r2 (see above), corresponding to Adh gene; rp49 s2 and rp49 as2 (see above), corresponding to rp49 gene; 5′-CGGCGAGGGGGGAAAAGGAC-3′ and 5′-CTTGGCAGCAGGTGGAAAATGTT-3′, corresponding to the 60D intergenic spacer.
RESULTS
The presence of rasiRNAs corresponding to a wide range of transposable elements requires Spn-E function
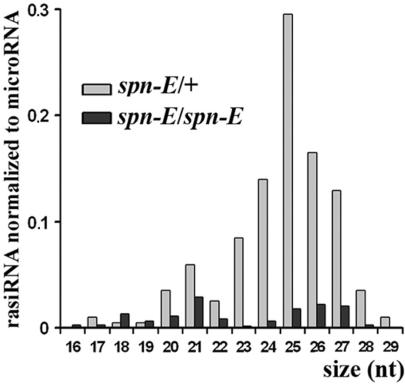
The spn-E (spindle-E, homeless) gene encodes a putative DExH box RNA helicase, which is required for rasiRNA-mediated silencing of selfish elements (28–30). Previously it was shown that the spn-E1 mutation leads to the loss of testis short RNAs related to the Su(Ste) repeats (2) and ovarian short RNAs of the HeT-A LINE element (30) and roo LTR retrotransposon (3). To address the effect of the spn-E gene on total rasiRNA abundance, we cloned short RNAs from spn-E1 homo and heterozygous ovaries and testes (Supplementary Table 1). In ovaries the quantity of rasiRNAs was 5-fold higher than that of miRNAs. This is a drastically increased ratio compared to the one calculated previously for Drosophila embryos and adult flies (about 0.65 and 0.1, respectively) (1). In contrast to ovaries, approximately equal amounts of microRNAs and rasiRNAs were observed in testes. The amount of rasiRNAs cloned from homozygous spn-E1 ovaries was 6.7 and 3.3 times lower than in heterozygotes if normalized to microRNA or to the sum of cloned fragments of ribosomal and transfer RNA, respectively (Figure 1). Both sense and antisense rasiRNAs abundance was decreased in spn-E1 homozygous ovaries. spn-E1 exerted the most pronounced effects on the amount of rasiRNAs related to LINE elements (Doc, F-element, G2 and R1A1) and some LTR retrotransposons (GATE, gypsy6 and MAX-element). The total amount of LINE-related rasiRNAs normalized to miRNAs was 20-fold lower in homozygous spn-E1 ovaries, whereas only a 4-fold decrease of LTR retrotransposon rasiRNA abundance was revealed (Supplementary Table 1).
Figure 1.
The spn-E1 mutation leads to the decrease of the overall rasiRNA abundance in ovaries. Length distribution of cloned rasiRNA and the ratios of rasiRNA to microRNA amounts are indicated by bars.
Derepression of transposable elements in the germline correlates with opening of chromatin structure
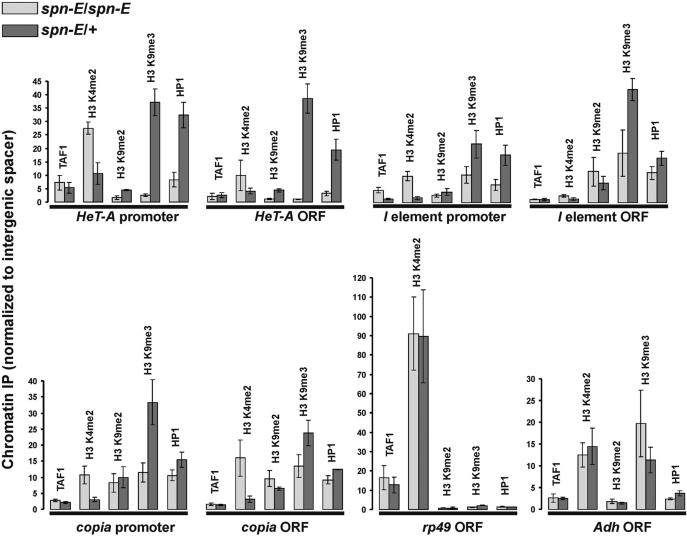
To investigate the role of chromatin state in rasiRNA-mediated transposable element silencing, we performed ChIP analysis of chromatin in ovarian nuclei using antibodies specific to known histone modifications. We focused on the three extensively investigated retrotransposons of Drosophila melanogaster: I element, HeT-A (LINE elements) and LTR-containing copia element. These three retrotransposons were shown to be up-regulated due to spn-E1 and other mutations, affecting the rasiRNA pathway in flies (3,28,30).
In spn-E1/+ heterozygous ovaries the chromatin of promoter and coding regions of tested retrotransposons compared with that of the ORF of the ribosomal rp49 gene contained a significantly lower level of histone H3 dimethylated at lysine 4 (H3 K4me2), the principal mark of transcriptionally active chromatin (31) (Figure 2). On the contrary, chromatin of retrotransposons was enriched with H3 K9me2 and particularly with H3 K9me3 mark, which are specific for inactive chromatin (32,33) (Figure 2). In spn-E1 homozygous ovaries we observed an increase in H3 K4me2 and a decrease in H3 K9me3 in promoters, as well as in coding regions of retrotransposons, but not in the chromatin of rp49 and Adh genes (Figure 2). Since methylation of H3 K4 was shown to be a cotranscriptional process (34), the increase in H3 K4me2 in the chromatin of retrotransposon coding regions may be considered as a consequence of an elevated level of their transcription.
Figure 2.
ChIP analysis of retrotransposons in ovarian chromatin. TAF1 occupancy and histone modifications in the chromatin of spn-E1/spn-E1 (light bars) and spn-E1/+ ovaries (dark bars) were tested. DNA in precipitates was measured by quantitative real-time PCR using primers to promoter and coding regions (ORF) of LINEs (HeT-A and I element) and LTR retrotransposon copia. The level of H3 K4me2 histone modification typical of transcriptionally active chromatin is significantly lower in retrotransposons, than in ORF of the constitutive rp49 gene. The spn-E1 mutation increases the level of H3 K4me2 and decreases the level of H3 K9me3 and HP1 both in promoters and ORFs of retrotransposons, but not in chromatin of rp49 and Adh genes. The obtained data were normalized to the fragment of intergenic non-transcribed spacer, located in the 60D region. The amplified spacer fragment contains no binding sites for any known chromatin proteins and does not belong to any repeat that may be silenced.
Along with endogenous retroelements, we performed ChIP analysis of a transgenic construct containing the reporter lacZ gene driven by copia LTR (copiaLTR-lacZ) located on the X chromosome. We also observed an increase of H3 K4me2 occupancy in spn-E1 homozygous ovaries, but no decrease of the repressive H3 K9me2 and H3 K9me3 marks (Supplementary Figure 2). The absence of this latter effect may be attributed to the euchromatic location of the copiaLTR-lacZ transgene compared to the mainly heterochromatic locations of endogenous copia elements.
The level of TAF1 protein, which is a known component of RNA polymerase II transcription initiation complex TFIID (35), remained unchanged in spn-E1 homozygous ovaries in HeT-A and copia promoters. The TAF1 level was increased 3-fold in the I element promoter (Figure 2) and increased 2-fold in copiaLTR-lacZ transgenic construct (Supplementary Figure 2). These results allow us to propose that chromatin opening is unlikely to occur as a result of enrichment with basal transcription factors in promoter regions.
We detected a significant amount of heterochromatic protein HP1 in the chromatin of I element, HeT-A and copia retrotransposons in ovaries. spn-E1 caused reduction of HP1 content in retrotransposons, especially for HeT-A (Figure 2). The 4-fold decrease of HP1 level was observed in promoters and 6-fold decrease in coding regions of HeT-A. At the same time, mutations in the HP1-encoding gene, which are available only in heterozygous state, lead to a drastic accumulation of HeT-A transcripts (36,37). This indicates that even a 2-fold decrease of HP1 level is sufficient for HeT-A derepression and the observed loss of HP1 occupancy of HeT-A chromatin, owing to spn-E1, causes transcriptional activation.
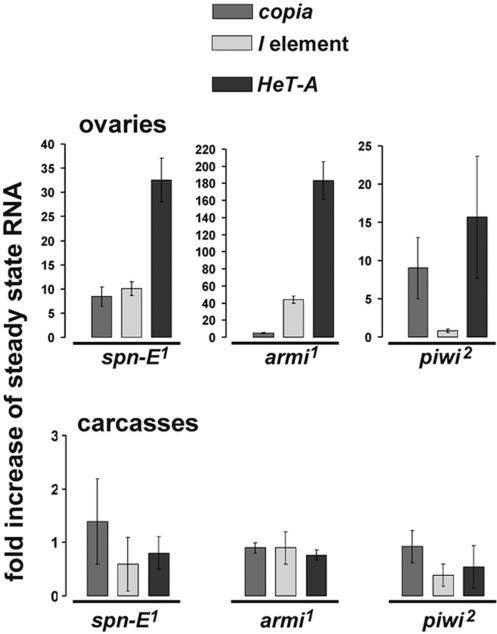
The most pronounced changes of the histone marks and HP1 level in HeT-A chromatin correlates with the most dramatic increase of HeT-A transcript level, compared to copia and I element in spn-E1 ovaries (Figure 3).
Figure 3.
Increase of retrotransposon transcript abundance in homozygous spn-E1, piwi2 and armi1 ovaries, but not in somatic tissues. The steady-state level of transcripts corresponding to HeT-A, I element and copia retrotransposons and house-keeping genes (rp49 and Adh) was detected by quantitative RT–PCR in ovaries and carcasses (flies without ovaries) of heterozygous or homozygous mutant females. Bars indicate the ratio of transcript amount in homozygous flies to heterozygous ones normalized to Adh transcript amount. piwi2 mutation produces less pronounced effects that may result from the severe morphological ovarian defects induced by this mutation.
rasiRNA-mediated chromatin silencing is restricted to the germline
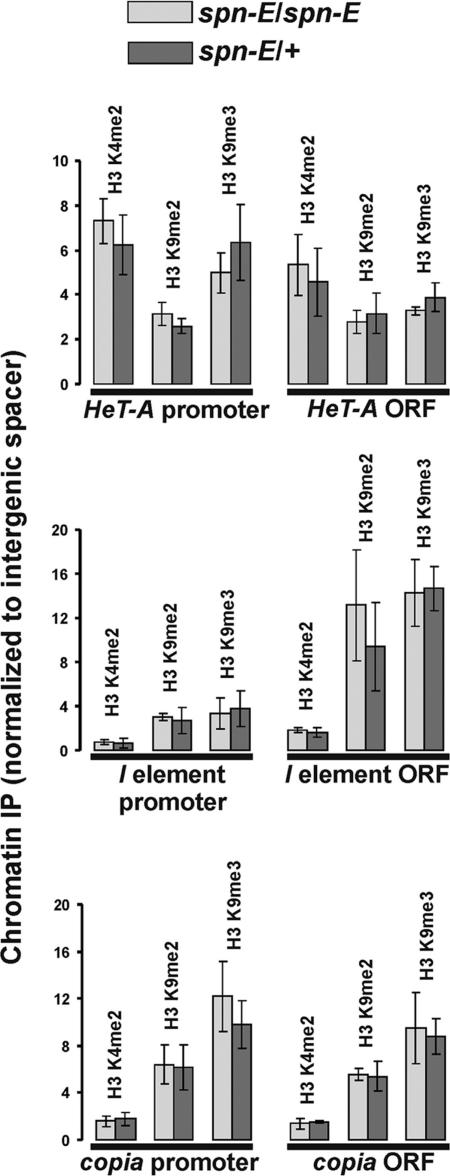
It was demonstrated that SPN-E, PIWI and AUB proteins are required for heterochromatin formation in somatic tissues of D. melanogaster (17). Some rasiRNA-pathway components have also been shown to be required for nuclear organization of a chromatin insulator (38), functioning of Polycomb chromatin complexes (39) and variegated repression of a white reporter carried by the 1360 element (40) in somatic tissues. The origin of short RNAs in these cases remains unknown. We investigated the regulation of Het-A, I element and copia in somatic tissues of flies, carrying mutations in the spn-E, piwi and armi genes, which control the rasiRNA pathway (3). The steady-state levels of HeT-A, I element and copia related to rp49 and Adh transcripts were comparable in ovaries, heads and carcasses (flies without ovaries) of spn-E1/+, piwi2/+ and armi1/+ heterozygous flies, indicating that tested retrotransposons are not exclusively germ-line transcribed. Nevertheless, we observed up-regulation of retrotransposon transcripts only in ovaries, but not in carcasses or heads of homozygous spn-E1, piwi2 and armi1 flies (Figure 3;data not shown). Furthermore, we found no effects of the spn-E1 mutation on the histone modifications in carcasses (Figure 4).
Figure 4.
ChIP assay in carcasses. The occupancy of histone modifications in the chromatin of retrotransposons in spn-E1/spn-E1 (light bars) and spn-E1/+ ovaries (dark bars).
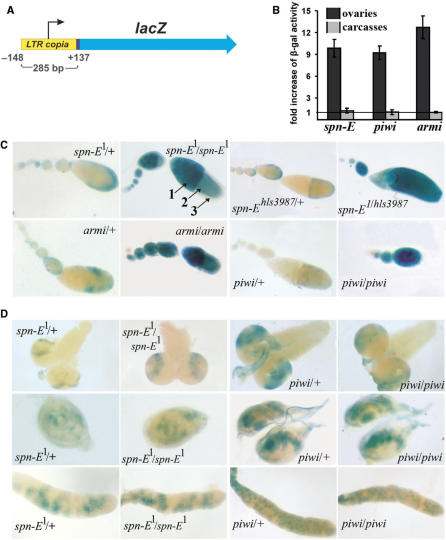
To extend the analysis of retrotransposon expression in somatic tissues, we used a transgenic copiaLTR-lacZ construct (Figure 5A). Activity of β-gal increased 10, 9 and 13 times in extracts of homozygous spn-E1/hls3987, piwi2 and armi1 ovaries, respectively, as compared to heterozygous ovaries, whereas the expression level remained unchanged in carcasses (Figure 5B). Expression of the construct was dramatically increased in germinal nurse cells and developing oocytes of spn-E1, piwi2, armi1 homozygous and spn-E1/hls3987 trans-heterozygous ovaries (Figure 5C). lacZ expression level remained unchanged in brain, imaginal discs and salivary glands of spn-E1, piwi2 and armi1 larvae as compared with heterozygous or wild-type controls (Figure 5D; data not shown). Thus, the chromatin-based regulation of tested retrotransposons mediated by rasiRNAs is realized in the germline.
Figure 5.
Structure and expression of the reporter construct copiaLTR-lacZ. (A) copia LTR comprises the known upstream regulatory region, including 137 bp of the transcribed fragment. (B) β-gal activity in ovaries and carcasses. Bars indicate the ratio of β-gal activity in spn-E1/hls3987 trans-heterozygous and piwi2, armi1 homozygous to heterozygous flies. The strong increase in β-gal activity due to the mutations was revealed in ovarian extracts (dark bars), whereas no significant changes were detected in extracts of carcasses (gray bars). Measurements of β-gal activity were normalized to total level of protein in extracts. (C) Expression of copiaLTR-lacZ located on the X-chromosome in spn-E1, piwi2, armi1 homozygous and trans-heterozygous spn-E1/hls3987 ovaries. In heterozygous spn-E1/+, spn-Ehls3987/+, piwi2/+ and armi1/+ ovaries lacZ expression occurs at a low level. In ovaries of homozygous and trans-heterozygous (spnE1/hls3987) females expression is increased in germinal nurse cells (arrow 2) and developing oocyte (arrow 1), but not in somatic follicle cells (arrow 3). (D) lacZ expression level is not increased in somatic organs of spn-E1 and piwi2 larvae. lacZ staining of larval brain (first row), haltere imaginal discs (second row) and salivary glands (third row) is shown.
DISCUSSION
We demonstrated that short rasiRNA species, known to be associated with Piwi subfamily proteins (3–6), have a germline-specific function in the maintenance of chromatin modifications of retrotransposons. The spn-E, piwi and armi genes are predominantly expressed in germ cells and their mutant states lead to abnormalities in germ-line development and sterility (41–44). Moreover, evidence of germ-line specificity of the rasiRNA-mediated silencing pathway is supported by the observation, that rasiRNAs are significantly more abundant in the germline than in somatic tissues. Germline-specific silencing of mobile elements is considered an important defense mechanism against mutations caused by mobile element transpositions, because selfish transposable elements are thought to be expressed mainly in germinal cells to ensure their amplification and transmission to the progeny. A distinct function of rasiRNA-mediated silencing concerns the maintenance of Drosophila telomeric state. Extension of telomeres is realized by germ-line specific transpositions of HeT-A, TAHRE and TART LINE elements (45–47). The aub and spn-E genes are implicated in the control of HeT-A and TART expression, accumulation of corresponding rasiRNAs and frequency of HeT-A and TART attachment to broken chromosome ends in ovaries (30). Thus, here we demonstrated the involvement of the rasiRNA pathway in the chromatin modification of beneficial telomeric retrotransposons and dangerous transposable elements.
We found that elimination of rasiRNAs in Drosophila ovaries caused by spn-E1 leads to the decompaction of chromatin of retrotransposons. The decrease of HP1 level and the changes in histone modification patterns, manifesting itself in an increase of H3 K4me2 and decrease of H3 K9me3 were observed. A correlation between the most dramatic increase of HeT-A transcript abundance, owing to the spn-E1, piwi2 and armi1 mutations (Figure 3), and the most significant changes of chromatin structure caused by spn-E1 compared to the I element, copia and copiaLTR-lacZ (Figure 2, Supplementary Figure 2) suggest that changes of chromatin structure in ovaries of rasiRNA mutants are accompanied by transcriptional activation of retrotransposons. At the same time, we detected no effects of spn-E1 on the chromatin state of retrotransposons in somatic tissues. The observed germline specificity of rasiRNA-mediated retrotransposon silencing is in apparent contradiction with the observations that the spn-E, piwi and aub mutations affect heterochromatin formation in somatic tissues (17,48) and these genes are required for variegated repression of a white reporter carried by the 1360 element (40). It is appropriate to point out that the size of the short RNAs corresponding to the 1360 element (40) and transgenic Fab-7 copies (39) (∼23 nt) in somatic tissues is consistent with Dicer-produced siRNAs, but not with rasiRNAs, suggesting that silencing of mobile elements in somatic tissues may be realized via RNAi, but not the rasiRNA pathway. Alternatively, rasiRNA-dependent heterochromatin formation might be induced in early stages of embryonic development and then be epigenetically inherited in somatic tissues in a rasiRNA-independent manner.
The mechanism of chromatin modification caused by rasiRNAs remains obscure. Although the Piwi protein was shown to be localized in cell nuclei (4,6,42), we failed to detect Piwi in the chromatin of retrotransposons (data not shown). Possibly, Piwi is associated with the nascent RNA but may easily leave chromatin. It has been suggested that rasiRNAs direct cleavage of retrotransposon transcripts (4–6). We propose that slicing of the nascent transcript mediated by the Piwi protein is capable to transform RNA polymerase II to a silencing complex. A similar model has been put forward to explain the spreading of transcriptional silencing in fission yeast Schizosaccharomyces pombe. It has been suggested that the sliced nascent transcript might recruit the silencing machinery to perform chromatin modification (49,50). Further experiments are required to verify this model of chromatin silencing in the D. melanogaster genome.
Our observations emphasize the proposed role of rasi(pi)RNAs in the formation of heterochromatin enriched by mobile elements and other repeats. Heterochromatin serves as a genome region to recruit and spread regulatory proteins to control chromosomal processes, including transcription as well as chromosome segregation. Actually, the disturbance of silencing of the Stellate repeats in the D. melanogaster genome is accompanied by chromosome meiotic non-disjunctions (51–53) and was shown to be triggered by spn-E mutations (22,51). Interestingly, spn-E mutations also lead to breakages in ovarian chromosomes (44) that might be caused by chromatin opening. The peculiarities of rasiRNA-dependent chromatin modification in Drosophila male and female germinal cells require further detailed studies taking into account the known role of heterochromatin in chromosome mechanics (54).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Alla Kalmykova for technical support in isolation of ovarian RNA used for short RNA cloning, Olesya Sokolova and Alina Korbut for help in RT-PCR and ChIP experiments, E.G. Pasyukova for kindly providing flies carrying the copiaLTR-lacZ construct and James M. Mason for help in the manuscript preparation. This work was supported by RAS program for Molecular and Cell Biology, Russian Foundation for Basic Research (05-04-48034), the program of Scientific School support (6113.2006.4) and grant of President of Russian Federation for young scientists (02.120.11.9326). Funding to pay the Open Access publication charges for this article was provided by Russian Foundation for Basic Research (05-04-48034).
Conflict of interest statement. None declared.
REFERENCES
- 1.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The Small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 2.Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 4.Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 6.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 7.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 8.Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 10.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 11.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 12.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 13.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- 15.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 16.Wassenegger M. The role of the RNAi machinery in heterochromatin formation. Cell. 2005;122:13–16. doi: 10.1016/j.cell.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 17.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 18.Gillespie DE, Berg CA. Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev. 1995;9:2495–2508. doi: 10.1101/gad.9.20.2495. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Reyes A, Elliott H, St Johnston D. Oocyte determination and the origin of polarity in Drosophila: the role of the spindle genes. Development. 1997;124:4927–4937. doi: 10.1242/dev.124.24.4927. [DOI] [PubMed] [Google Scholar]
- 20.Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 21.Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- 22.Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 23.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 24.Pfeffer S, Lagos-Quintana M, Tuschl T. Cloning of Small RNA Molecules. New York, NY: Wiley and Sons; 2003. [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chanas G, Lavrov S, Iral F, Cavalli G, Maschat F. Engrailed and polyhomeotic maintain posterior cell identity through cubitus-interruptus regulation. Dev. Biol. 2004;272:522–535. doi: 10.1016/j.ydbio.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 28.Vagin VV, Klenov MS, Kalmykova AI, Stolyarenko AD, Kotelnikov RN, Gvozdev VA. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 2004;1:54–58. [PubMed] [Google Scholar]
- 29.Kalmykova AI, Klenov MS, Gvozdev VA. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33:2052–2059. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 33.Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- 34.Eissenberg JC, Shilatifard A. Leaving a mark: the many footprints of the elongating RNA polymerase II. Curr. Opin. Genet. Dev. 2006;16:184–190. doi: 10.1016/j.gde.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 36.Savitsky M, Kravchuk O, Melnikova L, Georgiev P. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol. Cell Biol. 2002;22:3204–3218. doi: 10.1128/MCB.22.9.3204-3218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perrini B, Piacentini L, Fanti L, Altieri F, Chichiarelli S, Berloco M, Turano C, Ferraro A, Pimpinelli S. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol. Cell. 2004;15:467–476. doi: 10.1016/j.molcel.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 38.Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat. Genet. 2006;38:936–941. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- 39.Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 40.Haynes KA, Caudy AA, Collins L, Elgin SC. Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter. Curr. Biol. 2006;16:2222–2227. doi: 10.1016/j.cub.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cox DN, Chao A, Lin H. Piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 43.Williams RW, Rubin GM. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc. Natl Acad. Sci. USA. 2002;99:6889–6894. doi: 10.1073/pnas.072190799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 45.Biessmann H, Mason JM. Telomerase-independent mechanisms of telomere elongation. Cell Mol. Life Sci. 2003;60:2325–2333. doi: 10.1007/s00018-003-3247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melnikova L, Georgiev P. Drosophila telomeres: the non-telomerase alternative. Chromosome Res. 2005;13:431–441. doi: 10.1007/s10577-005-0992-7. [DOI] [PubMed] [Google Scholar]
- 47.Casacuberta E, Pardue ML. HeT-A and TART, two Drosophila retrotransposons with a bona fide role in chromosome structure for more than 60 million years. Cytogenet. Genome Res. 2005;110:152–159. doi: 10.1159/000084947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kavi HH, Fernandez HR, Xie W, Birchler JA. RNA silencing in Drosophila. FEBS Lett. 2005;579:5940–5949. doi: 10.1016/j.febslet.2005.08.069. [DOI] [PubMed] [Google Scholar]
- 49.Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- 50.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 51.Stapleton W, Das S, McKee BD. A role of the Drosophila homeless gene in repression of Stellate in male meiosis. Chromosoma. 2001;110:228–240. doi: 10.1007/s004120100136. [DOI] [PubMed] [Google Scholar]
- 52.Belloni M, Tritto P, Bozzetti MP, Palumbo G, Robbins LG. Does Stellate cause meiotic drive in Drosophila melanogaster? Genetics. 2002;161:1551–1559. doi: 10.1093/genetics/161.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boschi M, Belloni M, Robbins LG. Genetic evidence that nonhomologous disjunction and meiotic drive are properties of wild-type Drosophila melanogaster male meiosis. Genetics. 2006;172:305–316. doi: 10.1534/genetics.104.036806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dernburg AF, Sedat JW, Hawley RS. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]