Abstract
Toxic nitric oxide (NO) levels can regulate gene expression. Using a novel protein/DNA array, we show that toxic NO levels regulate the binding of trans-factors to various cis-elements in neuroblastoma cells, including CRE and those recognized by the transcription factors AP1, AP2, Brn-3a, EGR, E2F1 and SP1. Functionality of some of the cis-elements was confirmed by electro mobility shift and reporter assays. Interestingly, CREB, AP-1, Brn-3a, EGR and E2F1 can control mammalian cell viability. NO induced the anti-apoptotic Bcl-2 protein and its mRNA prior to the onset of death of 30–60% of the cells. Promoter analysis of the bcl-2 gene confirmed the involvement of a CRE in NO-dependent bcl-2 transcription. Neuroblastoma cells over-expressing bcl-2 became much more resistant to NO-induced apoptosis; conversely, Bcl-2 knockdown cells were rendered markedly more sensitive to NO. Together these results suggest that Bcl-2 counteracts NO-induced apoptosis in a fraction of the cell population. Thus, NO stimulates the binding of many trans-factors to their cognate cis-elements, some of which can regulate cell viability through transcriptional activation of target genes. Our results emphasize that a DNA/protein array approach can reveal novel, global transcription factor activities stimulated by cell death-regulating molecules.
INTRODUCTION
Nitric oxide (NO) is a ubiquitous signaling molecule synthesized from l-arginine in an NADPH- and O2-dependent reaction catalyzed by enzymes termed NO synthases (NOS) (1). NO is important for a variety of physiological functions such as vasodilation, fertilization, differentiation, inflammation and apoptosis (2–4). NO typically activates soluble guanylyl cyclase in cells to catalyze the conversion of GTP to cGMP that in turn activates cGMP-dependent protein kinase and other kinases, which mediate many of the normal physiological functions elicited by NO (5).
NO also influences cell viability either by inducing cell death when in excess or by protecting cells against various apoptotic or necrotic insults when present at more physiological levels (6,7). A predominant view is that excessive NO exerts cytotoxic effects in diverse cell types by reacting with superoxide and thereby generating the highly reactive free radical peroxynitrite, which causes non-specific oxidative DNA, protein and lipid damage. Such damage also triggers downstream signaling pathways and gene expression, which might elicit either cellular repair or apoptosis (6,8). A current view is that physiological NO concentrations can block apoptosis induced by various agents by activating anti-apoptotic/protective/repair proteins such as DNA-PK catalytic subunit, Bcl-XL, Bcl-2, cAMP-response element-binding protein (CREB), NAD(P)H Quinone oxidoreductase or heme oxygenase-1 (9–13). Conversely, excessive NO can elicit apoptosis or necrosis through a variety of mechanisms involving either p53-dependent or p53-independent pathways (14–16).
NO-dependent S-nitrosylation of proteins is increasingly being recognized as a key post-translational mechanism controlling the functions of certain proteins (17). There is accumulating evidence of selective S-nitrosylation of cysteines in proteins that can either block cell death or promote cell death; and many of these phenomena were observed in neurons (18,19). NO can regulate gene expression at least in part through the S-nitrosylation or phosphorylation of transcription factors or other proteins, and by regulating the cellular localization of transcription factors (17). Thus, S-nitrosylation contributes to the regulation of several pathways, including those leading to fos/Jun and NF-κB activation (20). Direct S-nitrosylation of the hypoxia-inducible transcription factor HIF-1α increases its DNA-binding capacity (17). NO induces phosphorylation of p53 that promotes its nuclear retention in neuroblastoma cells (21,22). The roles of NO in stimulating the functions of other transcription factors such as EGR-1 and Nrf2, but inhibiting the function of VDR/RXR, are documented (11,23–27). It is not clear if all these regulatory mechanisms (e.g. oxidative damage, S-nitrosylation, transcription) act in concert or independently, or function in a cell type-dependent manner. The actual outcome (cell death or repair) has been attributed to the particular concentrations of NO, the redox status of the cell and time-dependent differential gene regulation.
To reveal the cellular targets of NO under cell death promoting conditions, it is important to understand not only the specific S-nitrosylation events, but also the signaling pathways activated in response to NO. To this end, we used a novel protein/DNA array approach to determine the profiles of cis-elements/trans-factors that respond to apoptosis-inducing concentrations of NO in a neuronal cell line. We have identified a number of cis-elements that bind to various trans-factors in a NO-regulated fashion, analyzed the patterns of their regulation, and have begun the identification of the target genes they control in order to reveal the cascade of NO-dependent gene expression in the control of cell viability. We show that bcl-2 is a target gene of NO, and the consequent Bcl-2 up-regulation can limit the amount of apoptosis induced by toxic levels of NO.
MATERIALS AND METHODS
Materials
The human neuroblastoma cell line SH-Sy5y was obtained and grown as described previously (28). SNP, DETA-NO, SIN-1 and other chemicals were obtained from Sigma Aldrich. Except where stated, the enzymes used in this study were purchased from New England Biolabs. Platinum Taq polymerase, 10 mM dNTP mix and Lipofectamine transfection reagent were purchased from Invitrogen. The pcDNA-bcl-2-myc vector was a gift from Victor Yu, Institute of Molecular and Cell Biology, Singapore. The SureSilencing™ shBcl-2 Plasmid (targeting sequence in human Bcl-2: 5′-GAGGATTGTGGCCTTCTTTGA-3′) was purchased from SuperArray (KH00079N). The pGL3 Promoter vector containing the firefly luciferase gene, internal control plasmid pRL-TK that encodes Renilla luciferase, and the dual luciferase assay kit were from Promega (Madison, WI, USA). The Hybond ECL nitrocellulose membrane and ECL Western blot analysis kit were from Amersham Pharmacia Biotech. The RNeasy MiniKit was from Qiagen. The nuclear extraction kit was from Panomics Inc., CA, USA. The double-stranded oligonucleotides representing the cis-elements of AP1, AP2, BRN-3, CRE, EGR and SP1 and their mutants (Table 1), and the antibody against β-actin, were from Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA. The antibodies against Bcl-2 were from Upstate Biotechnology or Santa Cruz Biotechnology. The antibodies against caspase-3 were from Cell Signaling Technology or Santa Cruz Biotechnology. The Polyclonal antibody against GAPDH was from Abcam. The Transignal DNA/Protein Array I and Transcription Factor cDNA (TF cDNA) array were from Panomics Inc., CA, USA.
Table 1.
Oligonucleotide sequences of the various cis-elements and their mutant derivatives used in the construction of the reporter plasmids
| Cis-element | Oligonucleotide sequence |
|---|---|
| AP1-F | 5′ CTAG CGC TTG ATG ACT CAG CCG GAA 3′ |
| AP1-R | 5′ TCGA TTC CGG CTG AGT CAT CAA GCG 3′ |
| mAP1-F | 5′ CTAG CGC TTG ATG ACT TGG CCG GAA 3′ |
| mAP1-R | 5′ TCGA TTC CGG CCA AGT CAT CAA GCG 3′ |
| AP2-F | 5′ CTAG GAT CGA ACT GAC CGC CCG CGG CCC GT 3′ |
| AP2-R | 5′ TCGA AC GGG CCG CGG GCG GTC AGT TCG ATC 3′ |
| mAP2-F | 5′ CTAG GAT CGA ACT GAC CGC TTG CGG CCC GT 3′ |
| mAP2-R | 5′ TCGA AC GGG CCG CAA GCG GTC AGT TCG ATC 3′ |
| BRN-3-F | 5′ CTAG CAC AGC TCA TTA ACG CGC 3′ |
| BRN-3-R | 5′ TCGA GCG CGT TAA TGA GCT GTG 3′ |
| mBRN-3-F | 5′ CTAG CAC AGC TCA GCA ACG CGC 3′ |
| mBRN-3-R | 5′ TCGA GCG CGT TGC TGA GCT GTG 3′ |
| CRE-F | 5′ CTAG AGA GAT TGC CTG ACG TCA GAG AGC TAG 3′ |
| CRE-R | 5′ TCGA CTA GCT CTC TGA CGT CAG GCA ATC TCT 3′ |
| mCRE-F | 5′ CTAG AGA GAT TGC CTG TGG TCA GAG AGC TAG 3′ |
| mCRE-R | 5′ TCGA CTA GCT CTC TGA CCA CAG GCA ATC TCT 3′ |
| EGR-F | 5′ CTAG GGA TCC AGC GGG GGC GAG CGG GGG CGA 3′ |
| EGR-R | 5′ TCGA TCG CCC CCG CTC GCC CCC GCT GGA TCC 3′ |
| mEGR-F | 5′ CTAG GGA TCC AGC TAG GGC GAG CTA GGG CGA 3′ |
| mEGR-R | 5′ TCGA TCG CCC TAG CTC GCC CTA GCT GGA TCC 3′ |
| Sp1-F | 5′ CTAG ATT CGA TCG GGG CGG GGC GAG C 3′ |
| Sp1-R | 5′ TCGA G CTC GCC CCG CCC CGA TCG AAT 3′ |
| mSp1-F | 5′ CTAG ATT CGA TCG GTT CGG GGC GAG C 3′ |
| mSP1-R | 5′ TCGA G CTC GCC CCG AAC CGA TCG AAT 3′ |
The mutated core elements are shown in bold and italics.
Plasmid construction
The consensus oligonucleotide sequences/mutant sequences of AP1, AP2, BRN-3, CRE, EGR and SP1 as shown in Table 1 were subcloned in the pGL3 promoter vector to generate the reporter plasmids. The sense and antisense oligonucleotides corresponding to the cis-elements were synthesized with NheI and XhoI sites, respectively, and cloned at the respective sites in the pGL3 promoter vector. The reporter plasmids were named pGL3-AP1 Luc, pGL3-AP2 Luc, pGL3-BRN3 Luc, pGL3-CRE Luc, pGL3-EGR Luc and pGL3-SP1 Luc. The mutant reporter plasmids were named pGL3-mAP1 Luc, pGL3-mAP2 Luc, pGL3-mBRN3 Luc, pGL3-mCRE Luc, pGL3-mEGR Luc and pGL3-mSP1 Luc. The bcl-2 promoter constructs used are described (29).
Protein/DNA array analysis
SH-Sy5y cells were treated with NO donors for the times indicated, and nuclear proteins were prepared as described (11). The template of the protein/DNA array I and the details of the cis-elements present in the array are described previously (30). The protein/DNA array was performed following a procedure adapted from that described (30). Briefly, the nuclear extracts were incubated with biotin-labeled DNA probe mix in binding buffer for 30 min at 15°C. Protein/DNA complexes were resolved in a 2% agarose gel and excised. DNA probes were recovered from the protein/DNA complexes and hybridized to the transignal protein/DNA array membrane. Hybridization signals were visualized using horseradish peroxidase-mediated chemiluminescence. The experiment was repeated at least two times with two different membranes. The resulting autoradiograph was scanned by densitometry using a previously described method (30), and the fold induction/repression was tabulated along with the standard deviation (±SD) (Table 2).
Table 2.
NO-mediated induction or repression of trans-factor binding to cis-elements
| Cis-element | Fold Induction | Fold Repression |
|---|---|---|
| AP-1 | 1.98 ± 0.26 | – |
| AP-2 | 5.20 ± 1.67 | – |
| BRN-3 | 3.33 ± 0.59 | – |
| CBF | 3.75 ± 0.41 | – |
| CDP | 1.90 ± 0.30 | – |
| c-myb | 4.70 ± 1.40 | – |
| CRE | 5.83 ± 0.23 | – |
| E2F1 | 6.00 ± 1.93 | – |
| EGR | 3.25 ± 0.52 | – |
| Ets | 1.29 ± 1.10 | – |
| GAS/ | 1.95 ± 0.36 | – |
| GATA | 5.40 ± 0.42 | – |
| GRE | 2.90 ± 0.28 | – |
| MEF-2 | – | 4.52 ± 0.25 |
| NF-1 | 1.00 ± 0.08 | – |
| NF-E1 | 5.00 ± 0.33 | – |
| NF-E2 | 3.16 ± 0.15 | – |
| 1-Oct | 1.87 ± 0.04 | – |
| p53 | 1.32 ± 0.19 | - |
| Pax-5 | – | – |
| Pbx-1 | 3.50 ± 0.48 | – |
| EGR | 1.40 ± 0.25 | – |
| PPAR | – | 1.40 ± 0.30 |
| Sp1 | 3.02 ± 0.17 | – |
| TFIID | – | 4.10 ± 0.40 |
| TR | 4.07 ± 0.38 | – |
| USF-1 | 3.41 ± 0.74 | – |
The cis-elements that mediate significant induction or repression after treatment with the NO donor SNP (2 mM) for 4 h are tabulated. The fold of induction or repression was calculated by procedures given in the Materials and Methods section and represented along with the standard deviation (±SD).
Gel shift assays
SH-Sy5y cells were treated with the NO donors DETA-NO, SIN1 or SNP for various times up to 8 h. Nuclear extracts were prepared using the nuclear extraction kit from Panomics. The various cis-elements were end-labeled with [γ-32P] ATP and T4 polynucleotide kinase. Bandshift and the competition assays were performed by previously described procedures (31). Ten microgram of the nuclear extract was used in the gel shift assays.
Cell culture and transfection of reporter plasmids
The LipofectAMINE transfection reagent kit was used to transfect the various reporter plasmids in human SH-Sy5y neuroblastoma cells as described previously (28). The plasmid pRL-TK encoding Renilla luciferase was used as an internal control in each transfection. Thirty-six hours after transfection, the cells were washed three times with PBS and lysed in Passive Lysis buffer from The Dual-Luciferase Reporter Assay System Kit. The same kit was used to assay the samples for luciferase activity (31), which was measured with a TD-20e luminometer.
Transcription factor cDNA array analysis
SH-Sy5y cells were treated with NO donors for the indicated times, and RNA samples were prepared using a published method (11). The transcription factor array (TF array) was performed following the procedure described (30). Briefly, the probes were synthesized by combining 10 μg of total RNA (isolated from control and NO-treated SH-Sy5y cells) with 5 μl of TF cDNA primer mix provided by the manufacturer. The total mixture was heated for 2 min at 72°C followed by another 2 min at 42°C. The labeling mix contained 2 μl of Biotin-dUTP, 1 μl of reverse transcriptase and 12 μl of labeling mixture in water. The total labeling mix was mixed with the RNA-TF cDNA primer mix and incubated for 2 h at 42°C for the labeling reaction. After 2 h, 3 μl of the 10× denaturing solution was added to each sample and incubated for 20 min at 68°C. Finally, the probe was neutralized by adding 30 μl of 2× Neutralizing Buffer and incubating for 10 min at 72°C. Hybridization of the denatured probe to the transignal transcription factor cDNA array and subsequent washes were done according to the manufacturer's instructions. Hybridization signals were visualized using Streptavidin–HRP conjugate and horseradish peroxidase-mediated chemiluminescence. The experiment was repeated at least two times with two different membranes.
Western blot analysis
Western blot analysis was performed by methods as described (28). For total protein extraction, the cells were lysed in a buffer containing complete protease inhibitor cocktail. After centrifugation, 20 µg of total proteins were electrophoresed in 10% polyacrylamide gels and transferred to an ECL membrane. Immunoblotting was carried out with antibodies in phosphate-buffered saline with 0.2% Tween 20 and 5% BSA. After washing, the membrane was probed with horseradish peroxidase-conjugated donkey antiserum to rabbit or mouse (Chemicon) and developed by the enhanced chemiluminescence method (Amersham Pharmacia Biotech).
Semi-quantitative RT-PCR analysis
Total RNA was extracted from the cells after the NO donor treatment using the RNA extraction kit from Qiagen. Semi-quantitative RT-PCR analysis was performed using the one-step RT-PCR kit from Qiagen using the manufacturer's protocol. The primers used for the RT-PCR reactions were as follows. β-actin-Forward: 5′ atg gat gat gat atc gcc gcg ctc g 3′ and Reverse: 5′ gaa gca ttt gcg gtg gac gat gga ggg 3′; bcl-2-Forward: 5′ acc atggcgcac gctgggagaa cagg 3′ and Reverse 5′ cttgtggc ccagataggc acccaggg 3′; bclXL-2-Forward: 5′ acc atgtct cagagcaacc gggagctg 3′ and Reverse 5′ ttt ccgactgaag agtgagccca gcag 3′; bid-Forward: 5′ acc atggactgtg aggtcaacaa cgg 3′ and Reverse 5′ gtcca tcccatttct ggctaagctc c 3′; bad-Forward: 5′ acc atgttccaga tcccagagtt tgag 3′and Reverse 5′ ctgg gagggggcgg agcttcccct g 3′; fas-Forward: 5′ acc atgctg ggcatctgga ccctcc 3′ and Reverse 5′ gaccaagct ttggatttca tttctgaag 3′; fasL-Forward: 5′ acc atgcagcagc ccttcaatta cc 3′ and Reverse 5′ gag cttatataag ccgaaaaacg tctg 3′. In order to ensure the PCR reactions are in the exponential versus linear phase, we performed the PCR reactions for each target gene from 18 cycles up to 30 cycles, with 2 cycle intervals, and have carefully monitored the resulting products. The number of cycles up to which the product amplification is in the linear range for a particular target is taken into account for comparison. Accordingly, the cycling conditions used for the various primer sets were as follows. For β-actin, fas and fasL: 50°C for 30 min, 95°C for 15 min, followed by 26 cycles of 95°C for 1 min, 72°C for 2 min and a final extension at 72°C for 10 min. For bcl-2 and bcl-xL, the same reaction conditions as above were used, but the amplification was performed for only 22 cycles. For bad and bid: 50°C for 30 min, 95°C or 15 min, followed by 25 cycles of 95°C for 1 min, 62°C for 1 min, 72°C for 1 min and a final extension at 72°C for 10 min.
Stable cell selection, toxicity and enzymatic assays
To knock down Bcl-2, SH-Sy5y cells were stably transfected with the shBcl-2-containing plasmid using Lipofectamine™ 2000, and G418-resistant clones or vector control clones were isolated as described previously (32). To generate stable cell lines over-expressing Bcl-2, the plasmids pcDNA-Bcl-2-myc and the pcDNA vector were transfected individually in SH-Sy5y cells using LipofectAMINE 2000 following the manufacturer's protocol. The stable cells were selected in medium with 600 μg of neomycin/ml. After selection, the stable clones were maintained in medium containing 200 μg of neomycin/ml. To measure cell death, the Crystal Violet staining method, the caspase-3 activity assay or the LDH release assay was employed as described (32–34). For crystal violet staining, cells were plated in 6-well plates and either left untreated or treated with SNP, DETA-NO or SIN1 for 4, 8, 12, 16 and 24 h. The cells were stained with 30% Crystal Violet in 10% methanol for 10 min. Excess stain was removed completely by washing with water many times. The cells were dried, and the stain was eluted in 50% methanol, 1% acetic acid. Absorbance was measured at 590 nm (34). The activity of caspase-3-like proteases was measured using microtiter plates as described (32). The LDH release assay was done according to the published method (33). The statistical differences were determined using the Prism software by one-way analysis of variance (ANOVA) followed by the Tukey multiple comparison test.
RESULTS
NO induces apoptosis in SH-Sy5y cells
Previous work from our laboratory has demonstrated that NO induces apoptosis in a concentration and time-dependent manner in SH-Sy5y neuroblastoma cells (28,32). SH-Sy5y cells were treated with two different NO donors, SNP (2 mM) and DETA-NO (1.5 mM) for up to 24 h. Cell death was independently measured by Crystal Violet staining (Figure 1A) and caspase-3 activation (Figure 1B), confirming that NO donors (but not the vehicle control, left panel) induce substantial apoptotic death, the onset of which is notably delayed by at least 8 h.
Figure 1.
NO induces apoptosis in SH-Sy5y Cells. NO donor-induced cell death was quantitated using Crystal Violet staining or by measuring caspase-3 activity. (A) SH-Sy5y cells were untreated or treated with SNP (2 mM) or DETA-NO (1.5 mM) or media alone (control) and stained using Crystal Violet. The results are represented as percentage cell death. (B) Apoptosis was measured by caspase-3 activation in SH-Sy5y cells treated with SNP or DETA-NO. In each of panels A and B, the values represent mean ± SE of three independent experiments.
NO regulates binding of trans-factors to a collection of cis-elements
To identify the cis-elements whose binding to various trans-factors is regulated by NO, SH-Sy5y cells were treated with the NO donor SNP (2 mM) for 4 h. A novel protein/DNA array (Materials and Methods section) was used to identify the cis-elements whose binding to cognate trans-factors is increased or decreased by NO at 4 h. This time point was chosen, because it is prior to cell death when the cell is expected to be mounting transcriptional responses to the insult, but before the actual decision to survive or undergo apoptosis is taken. Table 2 shows that NO increased the binding of trans-factors to a collection of cis-elements from ∼1- to 6-fold. The maximum increase of 6-fold was observed with the E2F1 element, followed by elements such as GATA, AP2, NF-E1 and CRE (∼5-fold) (Figure 2A and Table 2). A moderate increase of ∼2- to 4-fold was observed with elements such as AP1, BRN-3, CBF, CDP, c-myb, NF-E2, PBX-1, EGR, GRE, SP1, TR and USF-1, whereas elements such as PPAR, MEF-2 and TFIID showed a moderate decrease in trans-factor binding (∼1- to 4-fold) (Figure 2A and Table 2). Elements such as SMAD and STAT did not show any increase or decrease in trans-factor binding with NO treatment (Figure 2A).
Figure 2.
NO induces the binding of trans-factors to an array of cis-elements. (A) SH-Sy5y cells were treated with SNP (2 mM) and the DNA/protein hybrid array (Panomics) was performed as described in Materials and Methods section. Detection of selected cis-elements is shown. (B and C) NO enhances the binding of nuclear factors to the various cis-elements. The cis-elements were end-labeled with γ-32P ATP. A total of 50 000 c.p.m. of the labeled cis-elements was incubated with 10 µg of nuclear extract from SH-Sy5y cells treated with the indicated NO donors for various times and analyzed in a 5% non-denaturing polyacrylamide gel. Shifted DNA–nuclear protein complexes are arrowed. (D) Competition assay. The wild-type (WT) cis-elements were end-labeled with γ-32P ATP. A total of 50 000 c.p.m. of labeled cis-elements were incubated with 10 µg of nuclear extract from SH-Sy5y cells treated with SNP either in the absence (control) or presence of 5×, 10× and 20× excess cold wild type (cold WT) or cold mutant (cold MT) oligonucleotide. The samples were analyzed in a 5% non-denaturing polyacrylamide gel. Shifted DNA–nuclear protein complexes are arrowed.
The increase in binding of the trans-factors to cis-elements observed with the protein/DNA hybrid array was subsequently confirmed by EMSA for some of the cis-elements (Figure 2B and C). We could not perform a confirmation for all the cis-elements that showed increases or decreases in binding in the array because of the large number of samples. For the EMSA, we used more than one NO donor (SIN1, SNP, DETA-NO) to confirm that the observed increase in binding of trans-factors to cis-elements is consistent irrespective of the donor compounds. The experiments were also done with different concentrations (0.5–2 mM) of NO donors at four different time points. All the NO donors increased the binding of trans-factors to the cis-elements AP1, AP2, CRE, EGR and SP1 very substantially (Figure 2B and C), and the magnitude of the increases was comparable to that observed with the protein/DNA array (Figure 2A and Table 2), though it was not identical. There was no consistent increase in the trans-factor binding to BRN-3. The competition assays with the cold (unlabeled) mutant oligonucleotides and the cold wild-type oligonucleotides showed that only the wild-type oligonucleotides could compete for binding (Figure 2D). This clearly shows the specificity of binding. Thus, the EMSA results confirm the NO-mediated increase in binding of trans-factors to many of the cis-elements observed with the protein/DNA array.
We next addressed whether the increase in the binding of trans-factors to cis-elements observed with the protein/DNA array and the EMSA translated into transcriptional activation or repression. Reporter plasmids carrying the luciferase gene linked to the various cis-elements or their mutated non-consensus sequences were constructed and used in transient transfection assays. Table 1 lists the oligonucleotides harboring the cis-elements and their mutants used in the construction of the reporter plasmids. The reporter plasmids pGL3-AP1 Luc, pGL3-AP2 Luc and pGL3-CRE Luc mediated a significant basal activity, which was strongly induced upon NO treatment (SNP 2 mM) in a time-dependent manner (Figure 3). The NO-induced activation observed with the various reporters after 12 h of NO treatment were ∼5-fold for AP1, ∼3-fold for AP2 and ∼12-fold for CRE (Figure 3, upper panels). The reporter plasmids pGL3-BRN3 Luc, pGL3-EGR Luc and pGL3-SP1 Luc also showed a substantial basal activity. However, the significant and sustained NO-mediated induction of transcriptional activity seen with the AP1, AP2 and CRE cis-element reporters upon NO treatment was not seen with BRN-3, EGR and SP1 (Figure 3, lower panels). The reporter plasmid pGL3-BRN3 Luc showed ∼1.5-fold activation 2 h after NO treatment, which reduced sharply after 2 h and reached the basal level by ∼8–12 h. The reporter plasmid pGL3-EGR Luc showed very marginal ∼0.2-fold activation by 2 h, but showed ∼1-fold repression by ∼8–12 h after NO treatment. The reporter plasmid pGL3-SP1 Luc did not show any significant activation upon NO treatment (Figure 3). The reporter assays were carried out at least three times with all three NO donors. The data presented in Figure 3 show the representative results obtained using SNP (2 mM) as the NO donor, although the results obtained from all three NO donors showed similar patterns. In pilot experiments with the transient transfections, 0.5–2.0 µg of the different reporter constructs were used, but the fold responses remained very similar (data not shown). This indicates a true result and not an artifact of transfecting excessive amounts of the reporter plasmids. Reporter assays with the plasmids carrying the mutant cis-elements did not exhibit any substantial basal activity, and the induction with the NO donors was completely abolished (data not shown). Hence, these results show that although NO activates the binding of transcription factors to a collection of cis-elements, not all these elements mediate transcriptional activation or repression.
Figure 3.
Effect of NO on transcriptional activation/repression mediated by the various cis-elements. Various cis-elements were cloned into the pGL3 vector with luciferase under the control of the SV40 promoter. SH-Sy5y cells were transfected with 0.5 µg of the various reporter plasmids along with 0.05 µg of the plasmid pRL-TK encoding Renilla luciferase. At 36 h post-transfection, the cells were treated with the NO donor SNP (2 mM) for the indicated times and analyzed for luciferase activity. The values represent mean ± SE of three independent transfection experiments.
Unchanged protein expression of transcription factors that bind to cis-elements
NO might induce or repress de novo transcription of the trans-factors that bind to the cis-elements, thereby contributing to the activation or repression observed with the protein/DNA array, EMSA and reporter assays. To address this issue, we employed a transcription factor cDNA array to measure the NO-regulated transcription factor expression levels. Overall, we did not observe any significant changes in the mRNA expression profiles of the clear majority of the transcription factors upon NO treatment using SNP (2 mM) as the NO donor (Figure 4). A slight increase in the NO-induced mRNA levels was observed with fosB and junB, whereas a decrease in mRNA levels was seen with junD and EGR2 (Figure 4). Thus, it cannot be ruled out that the NO-mediated reduction in EGR-Luc activity (Figure 3) may be due to lower expression of EGR2 induced by NO (Figure 4B). Nevertheless, we infer that NO mostly stimulates the binding of pre-formed transcription factors to various cis-elements, for example, by post-translational modifications (at the very least for AP-1, AP-2, CRE and Brn-3a), which consequently regulates gene transcription.
Figure 4.
Effect of NO on the expression of transcription factors. (A and B) SH-Sy5y cells were treated with the NO donor SNP (2 mM) for 4 and 8 h, and the transcription factor array was performed as described in the Materials and Methods section. The subsequent expression profiles of the selected transcription factors (representing mRNA levels) are shown.
The bcl-2 gene is a transcriptional target of NO
Is the NO-mediated transcriptional activation we observed mediated by the cis-elements physiologically relevant in terms of specific gene transcription? To identify the NO-regulated pro- and anti-apoptotic genes that harbor these cis-elements in their promoter regions, a semi-quantitative RT-PCR analysis was performed for the following genes—bid, bad, bcl-xL, fas, fasL and bcl-2 (these genes were selected for the analysis based on the fact that they have some of the cis-elements that we identified in our array in their promoter regions). No changes in the transcriptional profiles of bid, bcl-xL, (Figure 5) or bad, fas, fasL (data not shown) were observed with any of the NO donors. However, the transcript of the anti-apoptotic gene bcl-2 was significantly up-regulated as early as 2 h by all three NO donors (Figure 5A). Additional experiments done with IMR-32 neuroblastoma cells also showed a significant up-regulation of the bcl-2 gene as early as 2 h after treatment with the three NO donors (data not shown). An ∼2.5- to 4-fold induction of Bcl-2 protein was observed, which began as early as 2 h after SNP or DETA-NO treatment (Figure 5B). These experiments together suggest that the bcl-2 gene is a transcriptional target of NO.
Figure 5.
Bcl-2 gene is a transcriptional target of NO. (A) Semi-quantitative RT-PCR analysis by agarose gel electrophoresis. SH-Sy5y cells were treated with the NO donors DETA-NO, SNP or SIN1 in the indicated concentrations for various times, and semi-quantitative RT-PCR analysis was performed using 2 μg of total RNA for the bcl-2, bid and bcl-xL genes. β-actin is shown as a loading control. (B) Western blot analysis. SH-Sy5y cells were treated with 1.5 mM DETA-NO or 2 mM SNP for various times. Twenty microgram of total protein were loaded on a 10% polyacrylamide gel. The membrane was probed with a Bcl-2 monoclonal antibody (lower panels). The signals on the western blots were scanned, normalized against β-actin and represented as optical density to show the fold induction for bcl-2 at each time point (upper panels).
Cyclic AMP response element (CRE) mediates transcriptional regulation of bcl-2 by NO
The bcl-2 gene promoter comprises P1 and P2 regions and has characterized cis-elements viz., one CRE and two SP1 sites located upstream of the P1 promoter that determine the basal and inducible expression (Figure 6) (29). The various bcl-2 promoter constructs used in this study have been described (29) and are shown schematically in Figure 6A. SH-Sy5y cells were transfected with various full-length and truncated or mutated reporter plasmids. The full-length (P1 + P2) plasmid LB 322 harboring the bcl-2 gene promoter showed ∼1.5-fold inducible reporter activity with DETA-NO (1.5 mM) (Figure 6B). The construct LB 124 that comprises only the P1 region of the bcl-2 promoter retained the basal and NO-inducible activity of the full-length construct (LB 322: P1 plus P2). However, the construct LB 335, which has only the P2 region of the bcl-2 promoter, did not mediate any induction with NO (Figure 6B). In other words, the induction seen with the full-length construct (LB 322) upon NO treatment is solely from the P1 region and not from the P2 region of the bcl-2 promoter.
Figure 6.
cAMP response element (CRE) mediates transcriptional regulation of the bcl-2 gene by NO. (A) Schematic map of the human bcl-2 gene promoter constructs. The full-length human bcl-2 gene promoter harbors two well characterized regions, P1 and P2 (29). The LB 322 construct harbors the P1 and P2 regions upstream of the full-length promoter. LB 335 contains only the P2 region, whereas LB 124 contains only the P1 region. LB 334 contains a partial region of P1 harboring one CRE element and two SP1 elements. LB 595 is same as LB 334 with a mutation in the CRE site. LB 1263 and LB 1283 are same as LB 334 with LB 1263 harboring a mutation in the proximal SP1 site and LB 1283 harboring a mutation in the distal SP1 site, respectively. (B) Transcriptional regulation of the bcl-2 gene by NO is mediated by the P1 region only. SH-Sy5y cells were transfected with 0.5 µg of the various reporter plasmids and 0.05 µg of the plasmid pRL-TK encoding Renilla luciferase. At 36 h post-transfection, the cells were treated with DETA-NO (1.5 mM) for the indicated times and analyzed for luciferase activity. (C) Transcriptional regulation of bcl-2 gene by NO is mediated by a CRE element in the P1 region. SH-Sy5y cells were transfected with the various reporter plasmids, treated with DETA-NO (1.5 mM) and analyzed for luciferase activity as in B above. (D) Same as panel C, except that SNP (2 mM) was used instead of DETA-NO as NO donor. In each of panels B, C and D, the values represent mean ± SE of three independent transfection experiments. ▾1—distal Sp1 site, ▾2—CRE site, ▾3—proximal sp1 site, X—denotes mutation in the respective site.
To further characterize the cis-elements in the P1 region that respond to NO, a series of truncated and mutated reporter plasmids were used. The deletion construct LB 334 with two SP1 elements and one CRE element (Figure 6A) displayed similar basal and NO-inducible luciferase activity as that of the full-length (LB 322) and the P1 construct (LB 124) (Figure 6C). However, when the CRE element was mutated (LB 595), the basal activity was retained but the induction by DETA-NO (1.5 mM) was completely lost (Figure 6C). The constructs with a mutation in the proximal SP1 site (LB 1263) or the distal SP1 site (LB 1283) behaved in a similar manner to the full-length (LB 322) and truncated P1 (LB 124) promoter constructs (Figure 6C). Figure 6D shows that experiments with SNP (2 mM) as NO donor gave similar results as those in Figure 6C. These data strongly implicate the CRE element in NO-mediated bcl-2 gene induction.
Bcl-2 counteracts NO-induced apoptosis of SH-Sy5y cells
NO is known to induce apoptosis of SH-Sy5y cells in a time and concentration-dependent manner (Figure 1) (28,32). To study the functional relevance of CRE-mediated up-regulation of the bcl-2 gene in NO-induced apoptosis, stable cells expressing bcl-2 were generated, and two independent clones were selected and characterized (Figure 7). Two independent stable cell lines expressed higher levels of Bcl-2 protein as compared to the vector control cells (Figure 7A, left panel). The NO donors SNP (2 mM) and DETA-NO (1.5 mM) induced ∼30–70% cell death in the wild-type and vector control cells (Figure 7B), and caspase-3 activity was maximal by 12 h after NO treatment with SNP (2 mM) and DETA-NO (1.5 mM) (Figure 7C). In contrast, both independent stable cell lines expressing higher levels of Bcl-2 protein were substantially (60–70%) protected from NO-induced cell death (Figure 7B). In accord with this result, the bcl-2 stable cells exhibited a complete loss of caspase-3 activity, even at 16 h after NO stimulation (Figure 7C), and caspase-3 cleavage was strongly inhibited in these cells (Figure 7A, right panel).
Figure 7.
Stable cells over-expressing bcl-2 are highly resistant to NO-induced apoptosis. (A) SH-Sy5y cells were transfected with the plasmid pcDNA-Bcl-2-myc or the pcDNA vector to generate stable cells. Left panels, similar levels of expression of the bcl-2-myc over-expressed proteins (single arrows) in the stable clones (Clone #1, Clone #2) were visualized by western blot analysis. Right panels, western blot comparing NO-induced caspase-3 cleavage in the wild-type cells (WT), vector control (Vector) and the pcDNA-Bcl-2-myc stable clones (Clone #1, Clone #2). (B) SH-Sy5y wild-type cells (WT), vector control (Vector) and bcl-2 stable clones (#1, #2) were treated with the NO donors SNP (2 mM) or DETA-NO (1.5 mM) for the indicated times, and NO-induced cell death was quantitated using the LDH release assay. (C) SH-Sy5y wild-type cells (WT), vector control (Vector) and bcl-2 stable clones (#1, #2) were treated with the NO donors SNP (2 mM) or DETA-NO (1.5 mM) for the indicated times, and apoptosis was assessed by measuring caspase-3 activity.
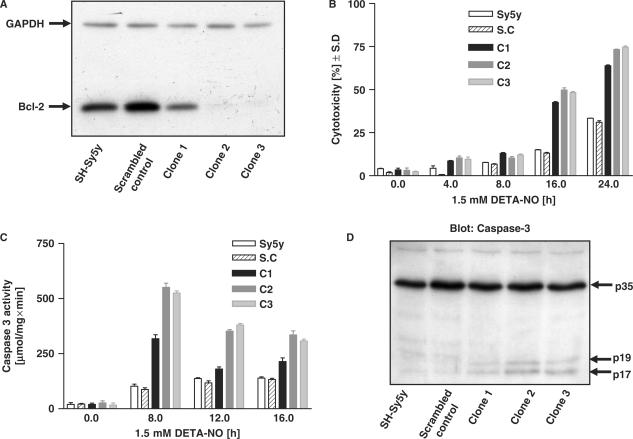
To corroborate these results and to address whether endogenous Bcl-2 can counteract NO-induced apoptosis, we knocked down Bcl-2 using a commercially validated shRNA (Materials and Methods section). Three individual Bcl-2 knockdown stable clones were generated, two of which almost totally abolished Bcl-2 synthesis (Figure 8A). In time course experiments, all three Bcl-2 knockdown clones exhibited increased NO-induced cell death and caspase-3 activity compared with the parental and vector control cells (Figure 8B and C). Consistent with Figure 8C, all three clones produced greater levels of caspase-3 cleaved fragments, indicative of caspase-3 activation (Figure 8D). Interestingly clone 1, which had the least Bcl-2 knockdown, showed levels of cytotoxicity, caspase-3 activity and caspase-3 cleaved fragments that were intermediate between the vector control and the fully knocked down clones 1 and 2.
Figure 8.
Stable knockdown of Bcl-2 sensitizes SH-Sy5y cells to NO-induced cell death. (A) Identification of stable Bcl-2 knockdown clones. Total proteins were subjected to electrophoresis in 0.1% SDS–14% polyacrylamide gels and western blot analysis for Bcl-2. Three individual clones (C1, C2 and C3) were analyzed. (B) SH-Sy5y parental cells, scrambled Bcl-2 oligonucleotide control cells (S.C) and three different clones of shBcl-2-expressing cells (C1–C3) were treated with 1.5 mM DETA-NO for the indicated times, and cytotoxicity was measured by LDH release. (C) As in B, except total proteins were harvested and caspase-3 activity was measured. (D) SH-Sy5y parental cells, vector control cells and three different clones of shBcl-2-expressing cells were treated with 1.5 mM DETA-NO for 16 h, and total proteins were subjected to electrophoresis as above followed by western blot analysis to detect procaspase-3 (p35) and cleaved caspase-3 fragments (p17, p19). Values in panels B and C are mean ± S.D determined from three experiments performed in triplicate.
Thus, the experiments in Figures 7 and 8 together illustrate the functional importance of NO-induced bcl-2 up-regulation and Bcl-2 synthesis (Figure 5) in counteracting NO-mediated apoptosis of neuroblastoma cells.
DISCUSSION
The plethora of responses elicited by toxic levels of NO may be determined by the particular reactive nitrogen intermediate, chemical modifications (e.g. S-nitrosylation) (17,18), or signal transduction pathways leading to transcriptional activation and gene expression that are increasingly being recognized as important aspects of the pleiotropic actions of NO (3,8,18). As will be discussed below, an emerging view is that excessive NO concentrations can simultaneously induce cell death promoting and counteracting signaling pathways, the balance and timing of which will determine whether the cell survives or dies.
The interactions of transcription factors with cis-acting regulatory elements may be considered the ultimate step in signal transduction pathways. Studies on NO-mediated transcriptional regulation have identified a very wide range of genes induced or repressed by NO in a cell-specific manner, anticipating that correspondingly diverse numbers of transcription factors may be involved in their expression or repression (3,8,16). In general, identification of transcription factors and their target genes has up till now been cumbersome; and not surprisingly, few studies have succeeded in addressing the actual cis-elements required for the physiological significance and target genes of the transcription factors activated by NO. Nevertheless, several transcription factors are known to be involved in NO-induced neuronal cell death or differentiation. These include c-Jun/AP-1 (bi-potential regulator of cell viability), CREB (which up-regulates protective Bcl-2), E2F (which down-regulates N-Myc in differentiation) and p53 (which up-regulates several pro-apoptotic genes)(6,7,28,32,35,36).
We used a novel protein/DNA array to investigate the cis-elements that display altered binding to cognate trans-factors in response to NO, identified a collection of cis-elements whose binding to trans-factors is regulated by NO, and analyzed their relevance to gene transcription in some cases. NO does not functionally activate trans-factor binding to all the known cis-elements; moreover, some cis-elements mediate gene activation, while others mediate gene repression, indicating that the effects that we observed are not non-specific. Most of the cis-elements in the array exhibited increased compared with decreased trans-factor binding, and some were confirmed by EMSA using different NO donors in varying concentrations. Although increased DNA binding was seen with some cis-elements, only a subset of them might have physiological relevance in terms of gene activation or repression as revealed by reporter assays.
Our transcription factor array analysis revealed CRE as one of the cis-elements whose binding to cognate trans-factors is strongly activated by NO, which confirmed our previous studies showing c-Jun/AP1 factors regulate NO-mediated cell survival and cell death in SH-Sy5y neuroblastoma cells (28,32), and also validates the whole array approach. In addition to c-Jun/AP1, the transcription factors AP-2, CREB, Brn-3a, Brn-3b and E2F have also been implicated in cell survival and cell death in various cell types (37–42). Hence, it is not surprising that we find that excessive NO activates the cis-elements that are bound by these transcription factors.
One of the novel and surprising findings in the present study is that NO induces a substantial ∼2- to 3-fold increase in the activity of AP2, a transcription factor mainly associated with the regulation of gene expression during development (43). AP2-regulated genes include those encoding p21WAF1/CIP1, c-Myc, TGF-α and c-Kit, which control cell division, differentiation or apoptosis (44–47). The cranial defects in the developing neuroepithelium and neural crest in AP-2α-deficient mice have been attributed, at least in part, to increased cell death (43). Moreover, AP-2 may act to attenuate c-Myc-dependent apoptosis under adverse conditions (44).
The NO-mediated activity profile of BRN-3 and EGR is unique in the sense that there is an initial moderate spike in trans-factor activity at the early time point (2 h) followed by a steady decrease in activity (from 4 to 12 h) (Figure 3). If indeed this translates to an in vivo effect in gene transcription it would be an interesting phenomenon of gene expression increasing in the early hours and decreasing steadily during the late hours following NO stress. In fact, this kind of gene profile has been reported for genes following various signaling events (3). Hence, it would be interesting to explore the downstream target genes harboring BRN-3 and EGR elements that respond to NO. In fact, some EGR- and Brn-3-regulated apoptotic genes are known, which provides a possible starting point (42,48,49). For instance, among the Brn-3 family of transcription factors, Brn-3a and Brn-3b are known to have different, sometimes antagonistic effects on specific promoters (41). Moreover, both Brn-3a and Brn-3b bind to p53 in neuroblastoma cells, but Brn-3a is associated with survival, growth arrest and differentiation, whereas Brn-3b enhances p53-mediated apoptosis (41). Another interesting finding in our study is the NO-mediated repression mediated through the cis-elements such as MEF-2, PPAR and TFIID. The cognate trans-factors for MEF-2 and TFIID were not known to be regulated by NO, and the significance of this repression by NO remains unknown (50,51). On the other hand, the response of the PPAR element to NO has been documented (52). More interestingly, the iNOS promoter has a functional PPAR-binding element (53). Hence a negative feedback regulation of NO synthesis by NO is possible through the PPAR element. However, this hypothesis needs to be verified experimentally. The data obtained with the SP1 reporters suggest that there is no obvious NO-regulated transactivation or transrepression. This finding suggests that although there is a strong increase in cis-element-trans-factor binding following NO treatment in the SP1 array and EMSA, it does not translate into an increase or decrease in transcriptional activity. The significance of the increase in DNA binding in the absence of any substantial change in transcriptional activity remains to be determined.
Finally, using the anti-apoptotic Bcl-2 protein as an example, we demonstrated that toxic concentrations of NO induce bcl-2 mRNA and subsequently its protein via a CRE element in the bcl-2 promoter. Interestingly, mutation in the CRE element did not affect the basal expression of the bcl-2 gene, implying that the CRE may not play any significant role in regulating the basal expression of the bcl-2 gene in SH-SY5Y cells (Figure 6). Cyclic AMP response element binding protein (CREB) is a major protein that binds to the CRE element. There is evidence that non-toxic levels of NO induce protection from apoptosis via CREB phosphorylation, CRE binding and consequent Bcl-2 up-regulation in neuronal cells (10,19,35). Lethal levels of NO donors might also activate gene expression via CREB (54); thus, it is entirely possible that CREB mediates NO-induced Bcl-2 up-regulation in the present study. Indeed, using lethal levels of NO donors, we showed Bcl-2 is up-regulated and trans-factor binding to CRE elements are both increased well before ∼30–60% cell death is evident. Even the amount of induced Bcl-2 is obviously not sufficient to prevent this (incomplete) cell death occurring in the parental cells, since over-expression of Bcl-2 totally prevents apoptosis. Conversely, complete knockdown of Bcl-2 not only sensitizes the cells to apoptosis at later time points, but also causes a much earlier activation of caspases—an indicator of the impending increased apoptosis. Although Bcl-2 is still induced at later time points when many of the cells (30–60%) are dying, we speculate this induction is occurring in the surviving cells.
Thus, we conclude Bcl-2 successfully counteracts NO-induced cell death in a significant fraction of neuroblastoma cells. In other words, it is likely that the up-regulation of Bcl-2 by NO both delays the onset of apoptosis and limits the number of cells that succumb to NO. Our findings are in accord with previous studies which hinted at a role for Bcl-2 by demonstrating a correlation between Bcl-2 levels and resistance of cells to NO-induced apoptosis (35,55,56). We have previously demonstrated that toxic NO levels stimulate the early transcriptional activation of the transcription factor Nrf2, which mitigates apoptosis of neuroblastoma cells via the synthesis of protective and anti-oxidant proteins (11). Thus, our functional analysis suggests that like Nrf2, NO-mediated Bcl-2 synthesis contributes to the overall cellular defense against excessive NO.
Overall our findings implicate a role for gene expression mediated by various cis-elements in delaying, counteracting or promoting apoptosis. Whether all the identified transcription factors and their cis-elements act in concert or independently needs to be investigated, though the failure of the majority of the tested transcription factors themselves to be regulated at the mRNA level by NO tends to argue that in the main, de novo transcription is induced or repressed by various preformed transcription factors downstream of various signal transduction pathways.
In summary, we show a DNA/protein array approach can readily reveal novel, global and functionally important transcription factor activities stimulated by signaling and toxic molecules or indeed any stimulus external to the cell. In general, the use of specific inhibitors of signaling proteins or knockout cells, combined with the array approach, will now allow us to determine whether particular signaling pathways are essential for regulating gene expression via one or more defined transcription factors.
ACKNOWLEDGEMENTS
We thank our colleagues for helpful comments, and Victor Yu for the pcDNA-bcl-2-myc vector. This work was supported by the Institute of Molecular and Cell Biology, Singapore. A.G.P. is an adjunct staff member of the Department of Surgery, National University of Singapore. Funding to pay the Open Access publication charges for this article was provided by the Institute of Molecular and Cell Biology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Michel T, Feron O. Nitric oxide synthases: which, where, how, and why? J. Clin. Invest. 1997;100:2146–2152. doi: 10.1172/JCI119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogdan C. The multiplex function of nitric oxide in (auto)immunity. J. Exp. Med. 1998;187:1361–1365. doi: 10.1084/jem.187.9.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemish J, Nakaya N, Mittal V, Enikolopov G. Nitric oxide activates diverse signaling pathways to regulate gene expression. J. Biol. Chem. 2003;278:42321–42329. doi: 10.1074/jbc.M308192200. [DOI] [PubMed] [Google Scholar]
- 4.Moncada S, Erusalimsky JD. Does nitric oxide modulate mitochondrial energy generation and apoptosis? Nat. Rev. Mol. Cell. Biol. 2002;3:214–220. doi: 10.1038/nrm762. [DOI] [PubMed] [Google Scholar]
- 5.Lucas KA, Pitari GM, Kazerounian S, Ruiz-Stewart I, Park J, Schulz S, Chepenik KP, Waldman SA. Guanylyl cyclases and signaling by cyclic GMP. Pharmacol. Rev. 2000;52:375–414. [PubMed] [Google Scholar]
- 6.Brune B, von Knethen A, Sandau KB. Nitric oxide (NO): an effector of apoptosis. Cell Death Differ. 1999;6:969–975. doi: 10.1038/sj.cdd.4400582. [DOI] [PubMed] [Google Scholar]
- 7.Li CQ, Wogan GN. Nitric oxide as a modulator of apoptosis. Cancer Lett. 2005;226:1–15. doi: 10.1016/j.canlet.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 8.Bogdan C. Nitric oxide and the regulation of gene expression. Trends Cell. Biol. 2001;11:66–75. doi: 10.1016/s0962-8924(00)01900-0. [DOI] [PubMed] [Google Scholar]
- 9.Choi BM, Pae HO, Chung HT. Nitric oxide priming protects nitric oxide-mediated apoptosis via heme oxygenase-1 induction. Free Radic Biol. Med. 2003;34:1136–1145. doi: 10.1016/s0891-5849(03)00064-9. [DOI] [PubMed] [Google Scholar]
- 10.Ciani E, Guidi S, Della VG, Perini G, Bartesaghi R, Contestabile A. Nitric oxide protects neuroblastoma cells from apoptosis induced by serum deprivation through cAMP-response element-binding protein (CREB) activation. J. Biol. Chem. 2002;277:49896–49902. doi: 10.1074/jbc.M206177200. [DOI] [PubMed] [Google Scholar]
- 11.Dhakshinamoorthy S, Porter AG. Nitric oxide-induced transcriptional up-regulation of protective genes by Nrf2 via the antioxidant response element counteracts apoptosis of neuroblastoma cells. J. Biol. Chem. 2004;279:20096–20107. doi: 10.1074/jbc.M312492200. [DOI] [PubMed] [Google Scholar]
- 12.Okada S, Zhang H, Hatano M, Tokuhisa T. A physiologic role of Bcl-xL induced in activated macrophages. J. Immunol. 1998;160:2590–2596. [PubMed] [Google Scholar]
- 13.Xu W, Liu L, Smith GC, Charles G. Nitric oxide upregulates expression of DNA-PKcs to protect cells from DNA-damaging anti-tumour agents. Nat. Cell. Biol. 2000;2:339–345. doi: 10.1038/35014028. [DOI] [PubMed] [Google Scholar]
- 14.Brune B. Nitric oxide: NO apoptosis or turning it ON? Cell Death Differ. 2003;10:864–869. doi: 10.1038/sj.cdd.4401261. [DOI] [PubMed] [Google Scholar]
- 15.McLaughlin LM, Demple B. Nitric oxide-induced apoptosis in lymphoblastoid and fibroblast cells dependent on the phosphorylation and activation of p53. Cancer Res. 2005;65:6097–6104. doi: 10.1158/0008-5472.CAN-04-4254. [DOI] [PubMed] [Google Scholar]
- 16.Li CQ, Robles AI, Hanigan CL, Hofseth LJ, Trudel LJ, Harris CC, Wogan GN. Apoptotic signaling pathways induced by nitric oxide in human lymphoblastoid cells expressing wild-type or mutant p53. Cancer Res. 2004;64:3022–3029. doi: 10.1158/0008-5472.can-03-1880. [DOI] [PubMed] [Google Scholar]
- 17.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell. Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 18.Benhar M, Stamler JS. A central role for S-nitrosylation in apoptosis. Nat. Cell Biol. 2005;7:645–646. doi: 10.1038/ncb0705-645. [DOI] [PubMed] [Google Scholar]
- 19.Contestabile A, Monti B, Contestabile A, Ciani E. Brain nitric oxide and its dual role in neurodegeneration/neuroprotection: understanding molecular mechanisms to devise drug approaches. Curr. Med. Chem. 2003;10:2147–2174. doi: 10.2174/0929867033456792. [DOI] [PubMed] [Google Scholar]
- 20.Marshall HE, Hess DT, Stamler JS. S-nitrosylation: physiological regulation of NF-kappaB. Proc. Natl Acad. Sci. USA. 2004;101:8841–8842. doi: 10.1073/pnas.0403034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneiderhan N, Budde A, Zhang Y, Brune B. Nitric oxide induces phosphorylation of p53 and impairs nuclear export. Oncogene. 2003;22:2857–2868. doi: 10.1038/sj.onc.1206431. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Zalcenstein A, Oren M. Nitric oxide promotes p53 nuclear retention and sensitizes neuroblastoma cells to apoptosis by ionizing radiation. Cell Death Differ. 2003;10:468–476. doi: 10.1038/sj.cdd.4401181. [DOI] [PubMed] [Google Scholar]
- 23.Berendji D, Kolb-Bachofen V, Zipfel PF, Skerka C, Carlberg C, Kroncke KD. Zinc finger transcription factors as molecular targets for nitric oxide-mediated immunosuppression: inhibition of IL-2 gene expression in murine lymphocytes. Mol. Med. 1999;5:721–730. [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura H, Weisz A, Kurashima Y, Hashimoto K, Ogura T, D'Acquisto F, Addeo R, Makuuchi M, Esumi H. Hypoxia response element of the human vascular endothelial growth factor gene mediates transcriptional regulation by nitric oxide: control of hypoxia-inducible factor-1 activity by nitric oxide. Blood. 2000;95:189–197. [PubMed] [Google Scholar]
- 25.Kroncke KD, Carlberg C. Inactivation of zinc finger transcription factors provides a mechanism for a gene regulatory role of nitric oxide. FASEB J. 2000;14:166–173. doi: 10.1096/fasebj.14.1.166. [DOI] [PubMed] [Google Scholar]
- 26.Matthews JR, Botting CH, Panico M, Morris HR, Hay RT. Inhibition of NF-kappaB DNA binding by nitric oxide. Nucleic Acids Res. 1996;24:2236–2242. doi: 10.1093/nar/24.12.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pilz RB, Suhasini M, Idriss S, Meinkoth JL, Boss GR. Nitric oxide and cGMP analogs activate transcription from AP-1-responsive promoters in mammalian cells. FASEB J. 1995;9:552–558. doi: 10.1096/fasebj.9.7.7737465. [DOI] [PubMed] [Google Scholar]
- 28.Feng Z, Li L, Ng PY, Porter AG. Neuronal differentiation and protection from nitric oxide-induced apoptosis require c-Jun-dependent expression of NCAM140. Mol. Cell. Biol. 2002;22:5357–5366. doi: 10.1128/MCB.22.15.5357-5366.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heckman CA, Mehew JW, Boxer LM. NF-kappaB activates Bcl-2 expression in t(14;18) lymphoma cells. Oncogene. 2002;21:3898–3908. doi: 10.1038/sj.onc.1205483. [DOI] [PubMed] [Google Scholar]
- 30.Jiang X, Norman M, Roth L, Li X. Protein-DNA array-based identification of transcription factor activities regulated by interaction with the glucocorticoid receptor. J. Biol. Chem. 2004;279:38480–38485. doi: 10.1074/jbc.M403948200. [DOI] [PubMed] [Google Scholar]
- 31.Dhakshinamoorthy S, Jaiswal AK. Small maf (MafG and MafK) proteins negatively regulate antioxidant response element-mediated expression and antioxidant induction of the NAD(P)H:Quinone oxidoreductase1 gene. J. Biol. Chem. 2000;275:40134–40141. doi: 10.1074/jbc.M003531200. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Feng Z, Porter AG. JNK-dependent phosphorylation of c-Jun on serine 63 mediates nitric oxide-induced apoptosis of neuroblastoma cells. J. Biol. Chem. 2004;279:4058–4065. doi: 10.1074/jbc.M310415200. [DOI] [PubMed] [Google Scholar]
- 33.Hentze H, Lin XY, Choi MS, Porter AG. Critical role for cathepsin B in mediating caspase-1-dependent interleukin-18 maturation and caspase-1-independent necrosis triggered by the microbial toxin nigericin. Cell Death Differ. 2003;10:956–968. doi: 10.1038/sj.cdd.4401264. [DOI] [PubMed] [Google Scholar]
- 34.Tosetti F, Vene R, Arena G, Morini M, Minghelli S, Noonan DM, Albini A. N-(4-hydroxyphenyl) retinamide inhibits retinoblastoma growth through reactive oxygen species-mediated cell death. Mol. Pharmacol. 2003;63:565–573. doi: 10.1124/mol.63.3.565. [DOI] [PubMed] [Google Scholar]
- 35.Ciani E, Guidi S, Bartesaghi R, Contestabile A. Nitric oxide regulates cGMP-dependent cAMP-responsive element binding protein phosphorylation and Bcl-2 expression in cerebellar neurons: implication for a survival role of nitric oxide. J. Neurochem. 2002;82:1282–1289. doi: 10.1046/j.1471-4159.2002.01080.x. [DOI] [PubMed] [Google Scholar]
- 36.Ciani E, Severi S, Contestabile A, Bartesaghi R, Contestabile A. Nitric oxide negatively regulates proliferation and promotes neuronal differentiation through N-Myc downregulation. J. Cell. Sci. 2004;117:4727–4737. doi: 10.1242/jcs.01348. [DOI] [PubMed] [Google Scholar]
- 37.Chiarugi V, Del Rosso M, Magnelli L. Brn-3a, a neuronal transcription factor of the POU gene family: indications for its involvement in cancer and angiogenesis. Mol. Biotechnol. 2002;22:123–127. doi: 10.1385/MB:22:2:123. [DOI] [PubMed] [Google Scholar]
- 38.Crosby ME, Almasan A. Opposing roles of E2Fs in cell proliferation and death. Cancer Biol. Ther. 2004;3:1208–1211. doi: 10.4161/cbt.3.12.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moser M, Pscherer A, Roth C, Becker J, Mucher G, Zerres K, Dixkens C, Weis J, Guay-Woodford L, et al. Enhanced apoptotic cell death of renal epithelial cells in mice lacking transcription factor AP-2beta. Genes Dev. 1997;11:1938–1948. doi: 10.1101/gad.11.15.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pilz RB, Casteel DE. Regulation of gene expression by cyclic GMP. Circ. Res. 2003;93:1034–1046. doi: 10.1161/01.RES.0000103311.52853.48. [DOI] [PubMed] [Google Scholar]
- 41.Budhram-Mahadeo VS, Bowen S, Lee S, Perez-Sanchez C, Ensor E, Morris PJ, Latchman DS. Brn-3b enhances the pro-apoptotic effects of p53 but not its induction of cell cycle arrest by cooperating in trans-activation of bax expression. Nucleic Acids Res. 2006;34:6640–6652. doi: 10.1093/nar/gkl878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Budram-Mahadeo V, Morris PJ, Latchman DS. The Brn-3a transcription factor inhibits the pro-apoptotic effect of p53 and enhances cell cycle arrest by differentially regulating the activity of the p53 target genes encoding Bax and p21(CIP1/Waf1) Oncogene. 2002;21:6123–6131. doi: 10.1038/sj.onc.1205842. [DOI] [PubMed] [Google Scholar]
- 43.Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- 44.Gaubatz S, Imhof A, Dosch R, Werner O, Mitchell P, Buettner R, Eilers M. Transcriptional activation by Myc is under negative control by the transcription factor AP-2. EMBO J. 1995;14:1508–1519. doi: 10.1002/j.1460-2075.1995.tb07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang S, Jean D, Luca M, Tainsky MA, Bar-Eli M. Loss of AP-2 results in downregulation of c-KIT and enhancement of melanoma tumorigenicity and metastasis. EMBO J. 1998;17:4358–4369. doi: 10.1093/emboj/17.15.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wajapeyee N, Somasundaram K. Cell cycle arrest and apoptosis induction by activator protein 2alpha (AP-2alpha) and the role of p53 and p21WAF1/CIP1 in AP-2alpha-mediated growth inhibition. J. Biol. Chem. 2003;278:52093–52101. doi: 10.1074/jbc.M305624200. [DOI] [PubMed] [Google Scholar]
- 47.Wang D, Shin TH, Kudlow JE. Transcription factor AP-2 controls transcription of the human transforming growth factor-alpha gene. J. Biol. Chem. 1997;272:14244–14250. doi: 10.1074/jbc.272.22.14244. [DOI] [PubMed] [Google Scholar]
- 48.Fu M, Zhu X, Zhang J, Liang J, Lin Y, Zhao L, Ehrengruber MU, Chen YE. Egr-1 target genes in human endothelial cells identified by microarray analysis. Gene. 2003;315:33–41. doi: 10.1016/s0378-1119(03)00730-3. [DOI] [PubMed] [Google Scholar]
- 49.Hudson CD, Morris PJ, Latchman DS, Budhram-Mahadeo VS. Brn-3a transcription factor blocks p53-mediated activation of proapoptotic target genes Noxa and Bax in vitro and in vivo to determine cell fate. J. Biol. Chem. 2005;280:11851–11858. doi: 10.1074/jbc.M408679200. [DOI] [PubMed] [Google Scholar]
- 50.Chen BS, Hampsey M. Transcription activation: unveiling the essential nature of TFIID. Curr. Biol. 2002;12:R620–R622. doi: 10.1016/s0960-9822(02)01134-x. [DOI] [PubMed] [Google Scholar]
- 51.Martin JF, Schwarz JJ, Olson EN. Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc. Natl Acad. Sci. USA. 1993;90:5282–5286. doi: 10.1073/pnas.90.11.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Knethen A, Brune B. Activation of peroxisome proliferator-activated receptor gamma by nitric oxide in monocytes/macrophages down-regulates p47phox and attenuates the respiratory burst. J. Immunol. 2002;169:2619–2626. doi: 10.4049/jimmunol.169.5.2619. [DOI] [PubMed] [Google Scholar]
- 53.Crosby MB, Svenson J, Gilkeson GS, Nowling TK. A novel PPAR response element in the murine iNOS promoter. Mol. Immunol. 2005;42:1303–1310. doi: 10.1016/j.molimm.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 54.Park SW, Sung MW, Heo DS, Inoue H, Shim SH, Kim KH. Nitric oxide upregulates the cyclooxygenase-2 expression through the cAMP-response element in its promoter in several cancer cell lines. Oncogene. 2005;24:6689–6698. doi: 10.1038/sj.onc.1208816. [DOI] [PubMed] [Google Scholar]
- 55.Kitamura Y, Kamoshima W, Shimohama S, Nomura Y, Taniguchi T. Nitric oxide donor-induced p53-sensitive cell death is enhanced by Bcl-2 reduction in human neuroblastoma cells. Neurochem. Int. 1998;32:93–102. doi: 10.1016/s0197-0186(97)00029-6. [DOI] [PubMed] [Google Scholar]
- 56.Melkova Z, Lee SB, Rodriguez D, Esteban M. Bcl-2 prevents nitric oxide-mediated apoptosis and poly(ADP-ribose) polymerase cleavage. FEBS Lett. 1997;403:273–278. doi: 10.1016/s0014-5793(97)00065-3. [DOI] [PubMed] [Google Scholar]