Abstract
Acanthocystis turfacea chlorella virus (ATCV-1), a prospective member of the family Phycodnaviridae, genus Chlorovirus, infects a unicellular, eukaryotic, chlorella-like green alga, Chlorella SAG 3.83, that is a symbiont in the heliozoon A. turfacea. The 288,047-bp ATCV-1 genome is the first virus to be sequenced that infects Chlorella SAG 3.83. ATCV-1 contains 329 putative protein-encoding and 11 tRNA-encoding genes. The protein-encoding genes are almost evenly distributed on both strands and intergenic space is minimal. Thirty-four percent of the viral gene products resemble entries in the public databases, including some that are unexpected for a virus. For example, these unique gene products include ribonucleoside-triphosphate reductase, dTDP-d-glucose 4,6 dehydratase, potassium ion transporter, aquaglyceroporin, and mucindesulfating sulfatase. Comparison of ATCV-1 protein-encoding genes with the prototype chlorella virus PBCV-1 indicates that about 80% of the ATCV-1 genes are present in PBCV-1.
Keywords: Chlorella viruses, Phycodnaviridae, Virus ATCV-1, Genome sequence, Acanthocystis turfacea
Introduction
Members and prospective members of the family Phycodnaviridae consist of a genetically diverse, but morphologically similar, group of large dsDNA-containing viruses (170–560 kb) that infect eukaryotic algae from both fresh and marine waters (Dunigan et al., 2006; Wilson et al., 2005b). The phycodnaviruses, together with the poxviruses, iridoviruses, asfarviruses, and the 1.2-Mb Mimivirus probably have a common evolutionary ancestor (Iyer et al., 2001, 2006; Raoult et al., 2004). All of these viruses share 9 gene products and at least two of these viral families encode an additional 32 homologous gene products (Iyer et al., 2006). Collectively, these viruses are referred to as Nucleo-Cytoplasmic Large DNA viruses (NCLDV) (Iyer et al., 2001).
Currently, the phycodnaviruses are grouped into 6 genera, based initially on host range and subsequently supported by sequence comparison of their DNA polymerases (Wilson et al., 2005b). Members of the genus Chlorovirus (chlorella viruses) infect fresh water algae, whereas members of the other five genera (Coccolithovirus, Phaeovirus, Prasinovirus, Prymnesiovirus, and Raphdovirus) infect marine algae. The genomes of prototype members from three of the Phycodnaviridae genera have been sequenced (Delaroque et al., 2001; Li et al., 1997; Wilson et al., 2005a). Comparative analysis of the three genomes have revealed more than 1000 unique gene products with only 14 gene products in common among the three genera (Dunigan et al., 2006). Thus the genetic diversity in the phycodnaviruses is enormous.
The chlorella viruses infect certain unicellular eukaryotic chlorella-like green algae that normally exist as endosymbionts in various protists, such as Paramecium bursaria (Kawakami and Kawakami, 1978; Van Etten et al., 1982) and Hydra viridis (Meints et al., 1981). P. bursaria chlorella virus (PBCV-1) is the type member of the group and has a 331-kb genome that was sequenced about 10 years ago. PBCV-1 has 366 putative protein-encoding genes and a polycistronic gene that encodes 11 tRNAs (Li et al., 1997).
To investigate the diversity of the chlorella viruses, we are sequencing the genomes of several additional family members. Previous reports describe the sequence and annotation of the 369-kb genome from virus NY-2A and the 345-kb genome from virus AR158, which, like PBCV-1, infect Chlorella NC64A (NC64A viruses) (Fitzgerald et al., in 2007b), and the 314-kb genome from virus MT325 and the 321-kb genome from virus FR483 that infect Chlorella Pbi (Pbi viruses) (Fitzgerald et al., 2007a). The current manuscript describes the sequence and annotation of the 288-kb genome from the recently discovered virus ATCV-1 (Bubeck and Pfitzner, 2005) that infects Chlorella SAG 3.83. Chlorella SAG 3.83 is normally a symbiont in the heliozoon Acanthocystis turfacea. Preliminary screening for plaque-forming viruses on Chlorella SAG 3.83 from fresh water samples collected in Germany, Canada, USA, Brazil, and England indicates that viruses that infect this host are common in nature (unpublished results).
Results and discussion
As part of the chlorella virus genome sequencing effort, a project Web site has been created at http://greengene.uml.edu. This site contains a database of the genomic DNA sequence assemblies as well as the predicted amino acid sequences of all virus-encoded open reading frames (ORF) and is viewable in text format or through a graphical genome browser. This database also contains the complete annotation for all chlorella virus-encoded ORFs. The supplemental data file referenced below is also available at this site.
Description of the viral genome
The ATCV-1 genome was assembled into a contiguous sequence of 288,047-bp, which agrees with the predicted size determined by pulse-field gel electrophoresis (unpublished results). Since the presumed hairpin termini were not sequenced, the left most nucleotide in the assembled sequences was designated nucleotide (nt) 1.
To orient the ATCV-1 genome relative to the sequenced NC64A and Pbi viruses, plots of proteins from the NC64A and Pbi virus prototype members, PBCV-1 and MT325 respectively, were compared with ATCV-1 proteins. These alignments reveal little co-linearity with the NC64A and the Pbi viruses (Fig. 1). Therefore, the genome was oriented by placing the 11 tRNA genes (see below) in the same direction as those encoded by PBCV-1 (i.e., transcribed from left to right). The tRNA genes in all of the chlorella virus genomes sequenced thus far are in this same orientation. The average G+C content of the ATCV-1 genome is 49.4%, a value higher than the ∼40% G + C content of the NC64A viruses and the ∼45% G+C content of the Pbi viruses (Fitzgerald et al.,2007a,b; Van Etten et al., 1985).
Fig. 1.
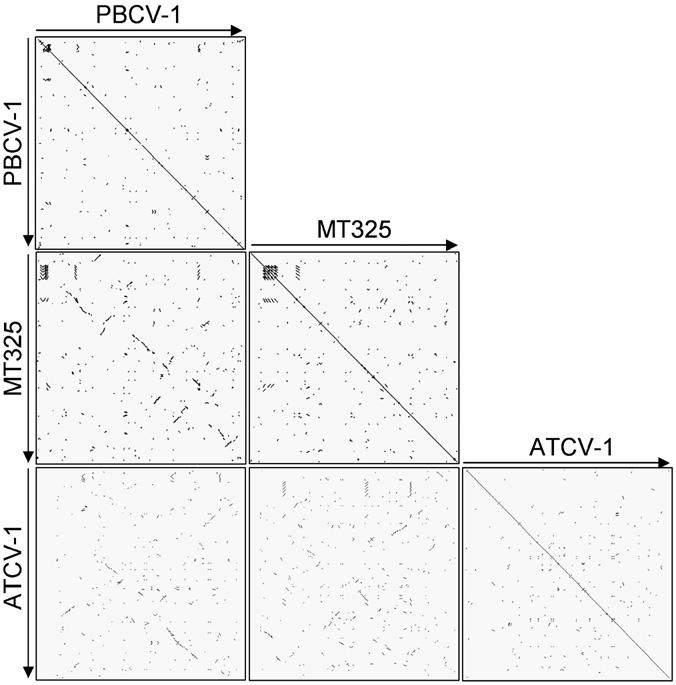
Comparison of three sequenced chlorella virus (PBCV-1, MT325, and ATCV-1) major ORFs with blastp dot plots. The dots represent ORF homology between two viruses with an E-value of less than 0.001.
Genes
A putative protein-encoding region, or ORF, was defined as a continuous stretch of DNA that translates into a polypeptide that is initiated by an ATG translation start codon and extends for 64 or more additional codons. The cut-off of 65 amino acid ORFs was first used in the annotation of the prototype PBCV-1 virus and has subsequently been used in the annotation of other chlorella viruses. Using this criterion, 860 ORFs were identified in the 288-kb ATCV-1 genome. The ORF names were based on three criteria. First, the ATCV-1 ORF names begin with either a “Z” for a major ORF (predicted to be a real protein-encoding gene) or a “z” for a minor ORF (not considered a true protein-encoding gene). Second, the ORFs were numbered consecutively in the order in which they appeared in the genome using the alignment with the PBCV-1 genome described above. Third, the letter R or L following the ORF number indicates that the transcript runs either left-to-right or right-to-left, respectively. The letters “Z” or “z” were chosen to name the ATCV-1 ORFs, which is the first virus sequenced that infects Chlorella SAG 3.83, to avoid confusion between the different chlorella viruses. The letters distinguish these viruses from those that infect Chlorella NC64A (i.e., PBCV-1, NY-2A, and AR158) or Chlorella Pbi (i.e., MT325 and FR483), designated with upper and lower case “A”, “B”, “C”, “M”, and “N”, respectively (Fitzgerald et al., 2007a,b).
The 860 ATCV-1 ORFs were classified into major or minor ORFs based on the following criteria. All of the ORFs were analyzed using the non-redundant, Pfam, and COG databases and ORFs predicted to encode a functional protein were classified as major. When an ORF classified as unknown based on sequence similarity to the databases, of either the same or opposite polarity, resided within or significantly overlapped another ORF, the larger ORF was classified as a major ORF and the smaller ORFs were classified as minor. These criteria led to the prediction that 329 of the 860 ATCV-1 ORFs probably encode proteins (Table 1).
Table 1.
Comparison of sequenced chlorella virus genomes
| Genome | General characteristics |
||||
|---|---|---|---|---|---|
| Host | Size (bp) | Genes | tRNA genes | G+C (%) | |
| PBCV-1 | NC64A | 330,743 | 366 | 11 | 40.0 |
| NY-2A | 368,683 | 404 | 7 | 40.7 | |
| AR158 | 344,690 | 360 | 6 | 40.7 | |
| MT325 | Pbi | 314,335 | 331 | 10 | 45.3 |
| FR483 | 321,240 | 335 | 9 | 44.6 | |
| ATCV-1 | SAG 3.83 | 288,047 | 329 | 11 | 49.4 |
The NC64A viruses have three types of introns in their protein-encoding genes. PBCV-1 and NY-2A have a self-splicing intron in a transcription factor TFIIS-like gene (Fitzgerald et al., 2007b; Li et al., 1997; Yamada et al., 1994). A splicesomal-processed intron is present in the DNA polymerase gene from all three sequenced NC64A viruses (Fitzgerald et al., 2007b; Grabherr et al., 1992; Zhang et al., 2001) and an 81-nt splicesomal processed intron exists in the pyrimidine dimer-specific glycosylase gene from some of the NC64A viruses (Fitzgerald et al., 2007b; Sun et al., 2000). The two Pbi viruses lack self-splicing and splicesomal-processed introns (Fitzgerald et al., 2007a). Analysis of the ATCV-1 genome indicates that its pyrimidine dimer-specific glycosylase gene (z313r) has an 83-nt phase 1 intron located at the same position as the 81-nt phase 1 intron in some of the NC64A viruses. However, the two introns have very little sequence identity. Also, like some of the other chlorella viruses (Fitzgerald et al., 2007a,b), one of the ATCV-1 tRNA genes is predicted to contain an intron (see below). No inteins were detected in ATCV-1.
GCG software was used to determine several general characteristics and properties for each ORF, including the nucleotide composition of the ORF, the A+T content of the 50 nts upstream of the ORF that is likely to contain the promoter region, the frame in which the putative protein was encoded, the number of amino acids in the encoded protein, the predicted protein molecular weight, and the predicted pI for each ORF. These properties are listed in Supplementary material 1. Some general characteristics for the ATCV-1 major ORFs are reported in Fig. 2, including the relative orientation of the ORFs (Fig. 2A). The directions in which the ORFs are encoded are slightly skewed in the reverse (∼55%) orientation. The average size of all the putative ATCV-1 proteins is 261 amino acids (Fig. 2B); about 50% of the proteins are 65 to 200 amino acids long. The predicted pIs of the proteins are depicted in Fig. 2C. Despite a trend for the proteins to have a pI in the 10–11 pH range, a peak also occurs at pH 4.5. Basic proteins have been identified in PBCV-1, NY-2A, and MT325 virions (Dunigan et al., manuscript in preparation). Therefore, some of the ATCV-1 basic proteins are probably associated with the virion where they presumably help neutralize the positively charged genomic DNA. However, the functions of the proteins that have pIs in the 4.5 range vary (e.g., ribonucleotide reductase small subunit, PCNA, and β-1,3-glucanase). Fig. 2D indicates the intergenic space between the major ORFs. Seventy-three percent of the major ORFs are separated by less than 100 nucleotides.
Fig. 2.
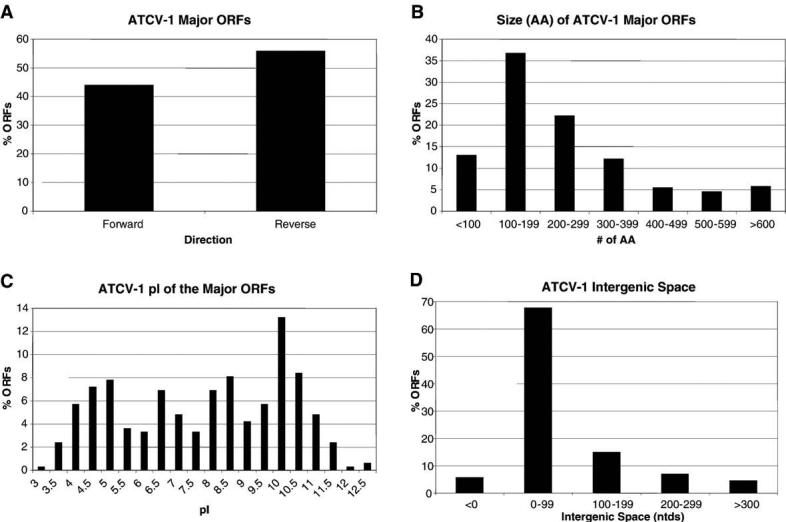
General characteristics of ATCV-1 major ORFs. (A) Orientation of the ORFs, (B) size of the ORFs, (C) predicted isoelectric points of the ORFs, and (D) intergenic space between the ORFs.
Annotation of the ATCV-1 genome
Every ORF was compared with the non-redundant database at NCBI using the criteria described in the Materials and methods section. The Pfam and the COG databases were used to identify conserved domains and proteins in the ATCV-1 ORFs (Supplementary material 1). A gene map of the ATCV-1 genome illustrates the location of the putative genes (Fig. 3) and some of the ORFs are listed by their predicted metabolic function (Table 2). No ATCV-1 gene products have been tested for activity. However, we assume that any ATCV-1-encoded proteins that have functional homologs in the other chlorella viruses are also functional.
Fig. 3.
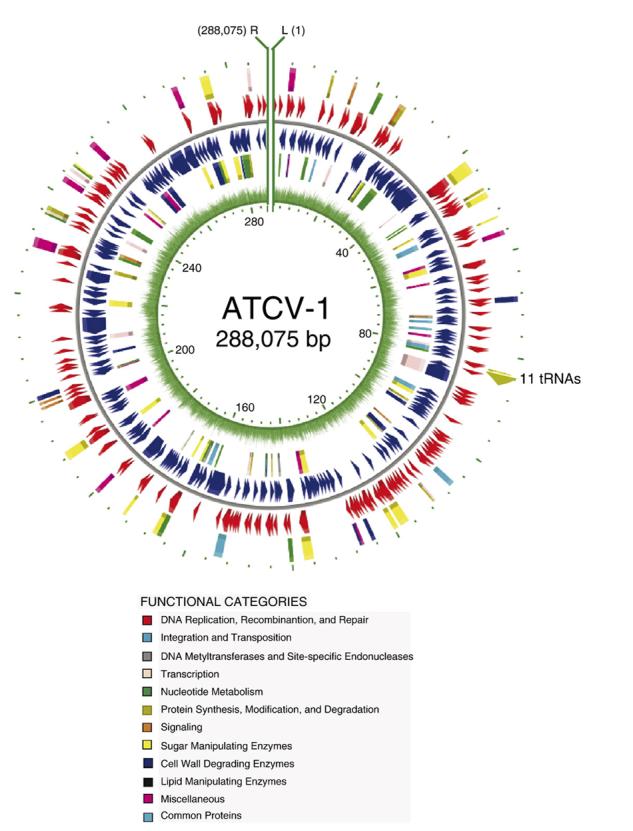
Map of the ATCV-1 genome arranged in a circle. However, the genome is a linear molecule and the ends are depicted at the top of the figure as green lines (L and R represent the left and right ends of the genome, respectively). The predicted ORFs are shown in both orientations and colored according to their functional category. The inner green circle represents the percent A + T across the genome using a 25 bp window.
Table 2.
ATCV-1 putative ORFs grouped by their functional categories
| DNA replication, recombination and repair | ||
|---|---|---|
| Description | ORF | AAa |
| δ DNA polymerase | Z798L | 903 |
| Archaeo-eukaryotic primase | Z054L | 425 |
| PCNA | Z788R | 293 |
| Z530L | 262 | |
| Replication factor C | Z453R | 378 |
| RNase H | Z356L | 167 |
| Helicase-Superfamily III | Z066R | 599 |
| DNA Topoisomerase II | Z551L | 1059 |
| ATP-dependent DNA ligase | Z187L | 301 |
| ATPase (PP-loop) | Z728L | 505 |
| ATPase (DNA packaging) | Z437R | 249 |
| Pyrimidine dimer-specific glycosylase | Z313R | 123 |
| Exonuclease | Z622R | 277 |
| Integration and tranposition | ||
| Description | ORF | AA |
| Tlr 6Fp DNA mobile protein | Z016R | 111 |
| GIY-YIG endonuclease | Z271R | 345 |
| Z311R | 247 | |
| Z378L | 300 | |
| Z387R | 255 | |
| Z490R | 203 | |
| Z548L | 295 | |
| Z633L | 235 | |
| Z637L | 234 | |
| Z641L | 253 | |
| Z737R | 155 | |
| HNH endonuclease | Z225R | 346 |
| Z350L | 358 | |
| Z441L | 210 | |
| Z578L | 240 | |
| Z614L | 100 | |
| Z692L | 368 | |
| Z782L | 367 | |
| Protein synthesis, modification and degradation | ||
| Description | ORF | AA |
| Translation elongation factor-3 | Z679L | 994 |
| Prolyl-4-hydroxylase | Z750R | 230 |
| Thiol oxidoreductase | Z061R | 117 |
| Protein disulfide isomerase | Z081L | 107 |
| SKP-1 protein | Z339L | 149 |
| Ubiquitin | Z203L | 78 |
| Ring finger ubiquitin ligase | Z292L | 229 |
| Ubiquitin C-terminal hydrolase | Z717R | 280 |
| Zn metallopeptidase | Z477L | 201 |
| Lipid manipulation | ||
| Description | ORF | AA |
| N-acetyltransferase | Z147L | 196 |
| Glycerophosphoryl diesterase | Z077L | 227 |
| Lipoprotein lipase | Z362R | 231 |
| Lysophospholipase | Z396R | 276 |
| Patatin-like phospholipase | Z612R | 272 |
| Signaling | ||
| Description | ORF | AA |
| Aquaglyceroporin | Z300R | 271 |
| Potassium channel protein | Z585R | 83 |
| Potassium ion transporter | Z696R | 645 |
| Dual specificity phosphatase | Z219L | 192 |
| Serine/Threonine protein kinase | Z575R | 569 |
| Z576R | 310 | |
| Z605R | 295 | |
| Z609R | 281 | |
| Z708L | 579 | |
| Cell wall degradation | ||
| Description | ORF | AA |
| Chitinase | Z780L | 467 |
| Z814L | 596 | |
| Chitosanase | Z204R | 341 |
| β & α 1,4 linked glucuronic lyase | Z771L | 358 |
| β-1,3-glucanase | Z819L | 318 |
| α-L-arabinofuranosidase | Z832L | 480 |
| DNA restriction/modification | ||
| Description | ORF | AA |
| Cytosine methyltransferase | Z040R | 350 |
| Nucleotide metabolism | ||
| Description | ORF | AA |
| Ribo. Reductase (small subunit) | Z035L | 326 |
| Ribo. Reductase (large subunit) | Z101L | 764 |
| Ribonucleoside-triphosphate reductase | Z838L | 628 |
| Deoxynucleoside kinase | Z457L | 189 |
| dCMP deaminase | Z616L | 150 |
| dUTP pyrophosphatase | Z500L | 163 |
| Thymidylate synthase X | Z818L | 243 |
| Glutaredoxin | Z134L | 109 |
| Z143R | 85 | |
| Thioredoxin | Z413L | 113 |
| Z476L | 149 | |
| Sugar manipulation | ||
| Description | ORF | AA |
| D-lactate dehydrogenase | Z295L | 347 |
| dTDP-D-glucose 4,6 dehydratase | Z544R | 351 |
| Mannose-6-phosphate isomerase | Z752L | 121 |
| GDP-D-mannose dehydratase | Z804L | 353 |
| Fucose synthase | Z282L | 320 |
| UDP-glucose 6-dehydrogenase | Z571L | 404 |
| Cellulase precursor | Z832L | 480 |
| Glycosyltransferase | Z120R | 846 |
| Z178L | 533 | |
| Z417L | 553 | |
| Z425R | 612 | |
| Z667L | 546 | |
| Z823R | 554 | |
| Transcription | ||
| Description | ORF | AA |
| Transcription factor TFIIB | Z716R | 289 |
| Transcription factor TFIID | Z502R | 286 |
| Transcription factor TFIIS | Z740R | 222 |
| VLTF2-type transcription factor | Z289L | 219 |
| Superfamily II helicase | Z257L | 1258 |
| Z596R | 708 | |
| Z643L | 448 | |
| mRNA guanylyltransferase | Z233L | 239 |
| RNA triphosphatase | Z084R | 200 |
| Histone H3, Lys 27 methylase | Z574L | 116 |
| SWI/SNF chromatin remodeling complex | Z794L | 1271 |
| SWI/SNF helicase | Z849R | 457 |
| RNase III | Z063L | 269 |
| Cytosine deaminase | Z744R | 123 |
| Miscellaneous | ||
| Description | ORF | AA |
| Ornithine/Arginine decarboxylase | Z760R | 372 |
| Agmatine iminohydrolase | Z806R | 363 |
| N-carbamoylput. amidohydrolase | Z169R | 299 |
| Homospermidine synthase | Z590L | 514 |
| Histidine decarboxylase | Z421L | 357 |
| Monoamine oxidase | Z773L | 387 |
| Amidase | Z177L | 279 |
| Cu/Zn-superoxide dismutase | Z190L | 183 |
| Erthrocyte binding protein | Z092L | 607 |
| ABC transporter protein | Z086R | 460 |
| Mucin-desulfating sulfatase | Z734R | 347 |
| Fibronectin binding protein | Z119L | 109 |
This number includes the stop codon so the actual amino acid number is one less than that listed.
Eighty-one percent of the ATCV-1 major ORFs are homologous to an ORF encoded by the prototype chlorella virus, PBCV-1. This finding suggests that the majority of major ORFs from the ATCV-1 virus are probably essential for virus replication in nature. The average amino acid identity between homologous proteins from PBCV-1 and ATCV-1 is 49%. ATCV-1 encodes the 9 gene products that are shared by all of the NCLD viruses (Iyer et al., 2001, 2006).
DNA replication and repair-associated proteins
ATCV-1 has 13 ORFs that are involved in either DNA replication, recombination, or repair, including δ-DNA polymerase (Z798L), DNA primase (Z054L), two sliding clamp processivity factor (PCNA) proteins (Z530L and Z788R), RNase H (Z356L), superfamily III helicase (Z066R), type II DNA topoisomerase (Z551L), ATP-dependent DNA ligase (Z187L), pyrimidine dimer-DNA glycosylase (Z313R), and exonuclease (Z622R) (Table 2). The ORF Z453R has been classified as a putative replication factor C due to its similarity to replication factor C's identified in other chlorella viruses. Some of these homologs, e.g., ORF A417L from PBCV-1, resemble putative replication factor C proteins from Plasmodium falciparum and P. yoelii; however, Z453R does not have much similarity to these proteins.
The ATCV-1 ATP-dependent type II DNA topoisomerase has approximately 40% amino acid identity with type II topoisomerases from several eukaryotic organisms. The Pbi virus CVM-1 encodes the smallest (1058 amino acids) characterized type II enzyme and it has DNA cleavage activity that is approximately 50-fold faster than the human type II topoisomerase (Dickey et al., 2005). The ATCV-1 type II DNA topoisomerase is the same size as its topoisomerase homologs from the Pbi viruses and has ∼71% amino acid identity.
PBCV-1 encodes the smallest functional ATP-dependent DNA ligase from a eukaryotic system (Ho et al., 1997) and the enzyme has been the subject of intensive mechanistic and structural studies (Sriskanda and Shuman, 2002 and references cited therein). The ATCV-1 ATP-dependent DNA ligase is similar in size to its homologs in the NC64A viruses (Fitzgerald et al., 2007b; Li et al., 1997); however, it only has ∼50% amino acid identity with the PBCV-1 enzyme. In contrast, the Pbi viruses lack a recognizable ATP-dependent DNA ligase.
Like the other chlorella viruses, ATCV-1 encodes two proteins that resemble PCNA proteins. The ATCV-1 proteins are more similar to their homologs from other organisms than they are to each other. This finding suggests that the viral PCNA genes did not arise by a recent gene duplication. PCNA interacts with proteins not only involved in DNA replication but also DNA repair and post-replicative processing, such as DNA methyltransferases and DNA transposases (Warbrick, 2000). Because the chlorella viruses encode proteins involved in both DNA repair and DNA methylation, the two PCNAs may serve different functions in their respective viral life cycles.
Transcription-associated proteins
No recognizable RNA polymerase components have been detected in any of the chlorella viruses that have been sequenced, including ATCV-1. This observation supports the concept that infectious viral DNAs are targeted to the nucleus and that host RNA polymerase(s) initiate(s) viral transcription, possibly in conjunction with virion-packaged transcription factors. ATCV-1 encodes at least four putative transcription factor-like elements: TFIIB (Z716R), TFIID (Z502R), TFIIS (Z740R), and VLTF2-type transcription factor (Z289L). However, none of these proteins is packaged in the PBCV-1 virion (Dunigan et al., manuscript in preparation) and is unlikely to be packaged in the ATCV-1 virion. ATCV-1 encodes two proteins that are involved in creating a mRNA cap structure, a mRNA guanylyltransferase (Z233L) and a RNA triphosphatase (Z084R). ATCV-1 also encodes a RNase III enzyme (Z063L) that is presumably involved in processing viral mRNAs and/or tRNAs.
ATCV-1, like all the other chlorella viruses, encodes two ORFs (Z596R and Z643L) that have superfamily II helicase domains. These helicases are involved in ATP-dependent DNA or RNA unwinding events that are needed in a variety of cellular processes. Except for the superfamily II helicase domains, the two proteins do not resemble one another. A third ATCV-1 ORF (Z257L) has homologs in all the chlorella viruses and some of these homologs have a conserved helicase C-terminal domain, but not Z257L.
In the immediate-early phase of infection, the host is reprogrammed to transcribe viral RNAs, which in PBCV-1 begins 5–10 min p.i. It is not known how this process occurs, but histone methylation may be involved in inhibiting host transcription. PBCV-1 encodes a 119-amino acid protein that contains a SET domain (named vSET) that di-methylates Lys27 in histone 3 (Manzur et al., 2003). vSET is packaged in the PBCV-1 virion and accumulating evidence indicates that vSET could be involved in repressing host transcription after PBCV-1 infection (Mujtaba et al., manuscript in preparation). ATCV-1 contains a vSET homolog (Z574L) that is slightly smaller (116 amino acids) than the PBCV-1 enzyme. The ATCV-1 enzyme has 55–66% amino acid identity to its vSET homologs in the five sequenced chlorella viruses. In addition to this histone methyltransferase, ATCV-1 encodes a putative SWI/SNF family helicase (Z849R) and a SWI/SNF chromatin remodeling complex protein (Z794L). Both proteins are also implicated in chromatin remodeling (Kim and Clark, 2002).
Finally, ATCV-1, as well as all the chlorella viruses, encodes a putative cytosine deaminase (Z744R). This observation suggests that either some of the viral transcripts or host transcripts may undergo some post-transcriptional editing (Gerber and Keller, 2001).
Protein synthesis, modification, and degradation
PBCV-1 was the first virus discovered to encode a translation elongation factor (EF) (Yamada et al., 1993). The PBCV-1 protein has about 45%-amino acid identity to an EF-3 protein from fungi (Belfield and Tuite, 1993; Chakraburtty, 2001). The fungal protein stimulates EF-1 GTP-dependent binding of amino acyl-tRNA to the ribosome A site. Like fungal EF-3 proteins, the virus-encoded proteins have an ABC transporter family signature and two ATP/GTP-binding site motifs. ATCV-1 has an ORF (Z679L) that encodes a 993 amino acid protein with 53–68% amino acid identity to the other chlorella virus EF-3 proteins.
ATCV-1 has several genes that encode proteins involved in post-translational modifications, including prolyl-4-hydroxylase (Z750R), protein kinases (see below), and glycosyltransferases (see below). ATCV-1 also encodes thiol oxidoreductase (Z061R), protein disulfide isomerase (Z081L), and a SKP-1 protein (Z339L). Additionally, the virus encodes three proteins involved in protein degradation, ubiquitin (Z203L), ubiquitin C-terminal hydrolase (Z717R), and ring finger ubiquitin ligase (Z292L). ATCV-1 also encodes a Zn metallopeptidase (Z477L).
tRNAs
ATCV-1 is predicted to encode 11 tRNAs: 2 for Asn and Val, and 1 each for Arg, Asp, Gly, Leu, Lys, Ser, and Tyr (Table 3). The two tRNAVal genes are 100% identical as are the two tRNAAsn genes suggesting that they are gene duplications. Unlike the central location of the tRNA genes in the Chlorella NC64A and Chlorella Pbi virus genomes, the ATCV-1 11 tRNA genes are clustered in a region a third of the way into the ATCV-1 genome, nucleotide sequence 83,727 to 84,858. Presumably, the tRNAs are transcribed as a large precursor RNA and processed via intermediates to mature tRNAs as they are in chlorella virus CVK2 (Nishida et al., 1999). Of the 11 ATCV-1 tRNAs, three are absent in the other sequenced chlorella viruses; tRNAAsp, tRNASer, and tRNAVal. There are 3 tRNAs which are present in each of the six sequenced chlorella viruses; tRNAArg, tRNAAsn, and tRNALeu. The remaining 5 tRNAs have homologs in at least two additional chlorella viruses. Although the orientation of the tRNA genes is the same in all six viruses, their order and location vary between the viruses. None of the tRNAs has a CCA sequence at the 3′ end of the acceptor stem. Typically, these three nucleotides are added post-transcriptionally.
Table 3.
ATCV-1 tRNA genes compared to tRNAs coded by other chlorella viruses
| tRNA | Anticodon | ATCV-1 |
PBCV-1 | NY-2A | AR158 | MT325 | FR483 | ||
|---|---|---|---|---|---|---|---|---|---|
| tRNA#a | Start | End | |||||||
| Ser | ACT | 1 | 83,727 | 83,793 | |||||
| Arg | TCT | 2 | 83,799 | 83,872 | + | + | + | + | + |
| Gly | TCC | 3 | 83,898 | 83,968 | + | + | |||
| Asp | GTC | 4 | 83,991 | 84,062 | |||||
| Val | AAC | 5 | 84,086 | 84,158 | + | + | + | ||
| Val | AAC | 6 | 84,181 | 84,253 | |||||
| Asn | GTT | 7 | 84,276 | 84,349 | + | + | + | + | + |
| Tyrb | GTA | 8 | 84,372 | 84,453 | + | + | + | + | |
| Lys | CTT | 9 | 84,456 | 84,528 | + | + | |||
| Asn | GTT | 10 | 84,551 | 84,624 | + | + | + | ||
| Leu | TAA | 11 | 84,773 | 84,858 | + | + | + | + | + |
+, tRNA gene present in virus.
Order of the tRNA genes in the ATCV-1 genome.
tRNA gene contains an intron.
One ATCV-1 tRNA, tRNATyr, is predicted to contain a 10-nt intron (Fig. 4A). The insertion of a 13-nt intron in the tRNATyr (anti-codon GTA) occurs in the same location in all but one sequenced chlorella virus genome, AR158 which lacks a tRNATyr (Fitzgerald et al., 2007a,b). The ATCV-1 tRNATyr intron has the highest identity (62%) to the intron from NY-2A and only has 38% and 31% identity to the introns from PBCV-1 and both Pbi viruses, respectively (Figs. 4B and C). Codon usage analyses of viral-encoded proteins indicate a strong correlation between the abundance of the virus-encoded tRNAs and their usage in viral proteins.
Fig. 4.
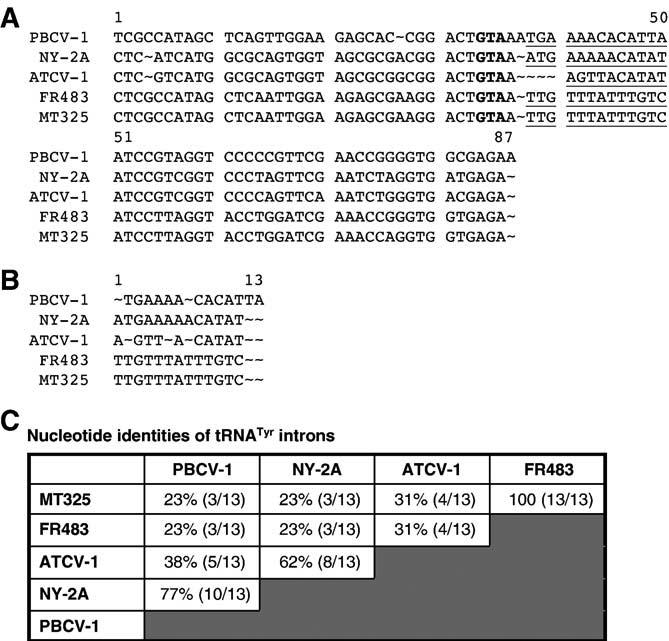
(A) Alignment of tRNATyr sequences from five sequenced chlorella virus genomes. The intron sequences are underlined; the “GTA” anticodon is in bold face type. (B) Alignment of tRNATyr intron sequences from five sequenced chlorella virus genomes. (C) Nucleotide identities between the tRNATyr intron sequences. Note that the percent identity is expressed out of 13 nts even though the ATCV-1 intron contains 10 nts.
Nucleotide metabolism
ATCV-1 encodes 11 enzymes involved in nucleotide metabolism. These enzymes are important because the DNA concentration in PBCV-1-infected cells increases at least four-fold following infection (Van Etten et al., 1984). Therefore, large quantities of dNTPs must be synthesized to support viral DNA replication. ATCV-1 encodes the small (Z035L) and large (Z101L) subunits of ribonucleotide reductase, deoxynucleoside kinase (Z457L), deoxycytidylate (dCMP) deaminase (Z616L), deoxyuridine triphosphate (dUTP) pyrophosphatase (Z500L), thymidylate synthase X (Z818L), two glutaredoxins (Z134L and Z143R; 35% amino acid identity with each other), and two thioredoxins (Z413L and Z476L; 28% amino acid identity to each other). ATCV-1 also encodes a ribonucleoside-triphosphate reductase (Z838L), which is the first time a gene encoding this enzyme has been found in the chlorella viruses. Ribonucleoside-triphosphate reductases are involved in purine and pyrimidine metabolism by converting 2′deoxyribonucleoside triphosphate and thioredoxin disulfide to ribonucleoside triphosphate and thioredoxin.
Two ATCV-1 enzymes, dUTP pyrophosphatase and dCMP deaminase, produce dUMP, the substrate for thymidylate synthetase. The chlorella viruses, including ATCV-1, lack a traditional thymidylate synthetase A. Instead, the viruses encode a protein that is a member of a newly recognized family of flavin-dependent thymidylate synthetases, named ThyX (Graziani et al., 2004; Myllykallio et al., 2002).
Protein kinases, phosphatases, and channel proteins
ATCV-1 encodes 5 Ser/Thr protein kinases (Table 2) and a protein that resembles a dual-specificity phosphatase (Z219L). The large number of virus-encoded proteins involved in phosphorylation/dephosphorylation suggests that they are involved in one or more signal transduction pathways that are important for virus replication.
The chlorella viruses were the first viruses to encode proteins that form K+ channels (called Kcv). Expression of kcv genes from 40 NC64A viruses and one Pbi virus, MT325, in Xenopus oocytes results in the formation of a functional K+ channel (Gazzarrini et al., 2006; Kang et al., 2004; Plugge et al., 2000). ATCV-1 is the first chlorella virus that codes for both a Kcv (Z585R) and a potassium ion transporter protein (Z696R). The ATCV-1 Kcv, which is 83 amino acids in size, is 11 amino acids smaller than the Kcvs from the NC64A viruses (94 amino acids). If ATCV-1 Kcv forms a functional channel in oocytes, it will be the smallest protein known to form a functional K+ channel. Preliminary studies indicate that ATCV-1 Kcv complements a yeast mutant lacking a potassium ion channel (Kang, unpublished results).
ATCV-1 and one other chlorella virus, MT325, encode an aquaglyceroporin channel (called AQPV, Z300R). The MT325 AQPV forms a functional channel in oocytes (Gazzarrini et al., 2006). The ATCV-1 AQPV is the same size as its MT325 homolog and the two proteins have 75% amino acid identity.
Sugar- and lipid-manipulating proteins
ATCV-1 has 13 genes that encode proteins with high identities to enzymes involved in either manipulating sugars, synthesizing polysaccharides, or transferring sugars to proteins. Two of the viral encoded enzymes, GDP-d-mannose dehydratase (GMD) (Z804L) and fucose synthase (Z282L), comprise a three-step pathway that converts GDP-d-mannose to GDP-l-fucose. The ATCV-1 GMD has 68% amino acid identity to GMDs found in bacteria, 62% amino acid identity to a cyanophage but only 52% amino acid identity to the PBCV-1 GMD. Unexpectedly, the PBCV-1 GMD differs from other GMDs because, in addition to the dehydratase activity, the protein also has a strong stereospecific NADPH-dependent reductase activity that produces GDP-d-rhamnose (Tonetti et al., 2003). It will be interesting to determine if the ATCV-1 GMD has two enzymatic activities like the PBCV-1 enzyme or if it only has one activity like the bacterial GMDs.
The chlorella viruses are also unusual because some of them encode enzymes involved in the biosynthesis of the linear polysaccharides hyaluronan (hyaluronan synthase) and/or chitin (chitin synthase) (DeAngelis et al., 1997; Graves et al., 1999; Kawasaki et al., 2002; Ali et al., 2005). Furthermore, some of the viruses encode enzymes involved in synthesizing the sugars that comprise the polysaccharides, e.g., UDP-glucose dehydrogenase and glutamine:fructose-6-phosphate amidotransferase (Landstein et al., 1998). These results led to the completely unexpected discovery that hyaluronan and/or chitin begin to accumulate on the surface of host cells shortly after infection (Graves et al., 1999; Kawasaki et al., 2002). Unlike many of the other chlorella viruses, ATCV-1 does not encode hyaluronan synthase or chitin synthase homologs. Furthermore, ATCV-1 is the first chlorella virus to lack a glutamine:fructose-6-phosphate amidotransferase encoding gene. However, ATCV-1 does encode an UDP-glucose dehydrogenase (Z571L). ATCV-1 also has genes encoding two sugar manipulating enzymes that are unique to this virus, mannose-6-phosphate isomerase (Z752L) and dTDP-d-glucose 4,6 dehydratase (Z544R). dTDP-d-glucose 4,6 dehydratase is the second enzyme in a four enzyme pathway that converts glucose-1-phosphate and dTTP to dTDP-1-rhamnose. The ATCV-1 120 amino acid mannose-6-phosphate isomerase appears to be truncated and may not be functional; it has ∼40% amino acid identity to the C-termini of mannose-6-phosphate isomerases (∼460 amino acids) from bacteria and archaea.
ATCV-1 encodes six putative glycosyltransferases (Table 2) which are probably involved in glycosylation of the virus major capsid protein (Graves et al., 2001). This is the most glycosyltransferases encoded by any chlorella virus sequenced to date.
ATCV-1 also encodes 5 enzymes involved in lipid metabolism including N-acetyl-transferase (Z147L), glycerophosphoryl diesterase (Z077L), lipoprotein lipase (Z362R), lysophospholipase (Z396R), and patatin phospholipase (Z612R).
Cell wall-degrading enzymes
ATCV-1 encodes six proteins that may be involved in degrading cell walls either during virus infection or virus release. These proteins include two chitinases (Z780L and Z814L), a chitosanase (Z204R), a β-1,3-glucanase (Z819L), and a polysaccharide lyase that cleaves chains of either β-or α-1,4 linked glucuronic acids (Z771L) (Sugimoto et al., 2004). All of the cell wall-degrading enzymes have functional homologs in PBCV-1 (Yamada et al., 2006). Finally, ATCV-1 encodes a protein (Z832L) that has an α-l-arabinofuranosidase domain and resembles some cellulases. Z832L homologs are also present in three of the other chlorella viruses, AR158, MT325, and FR483.
Restriction-modification enzymes
Chlorella viruses contain different levels of 5-methylcytosine (5 mC) and N6-methyladenine (6 mA) in their genomes (Van Etten et al., 1991). Therefore, it is not surprising that these viruses encode 5 mC and 6 mA DNA methyltransferases, e.g., NY-2A and PBCV-1 encode 18 and 5 DNA methyltransferases, respectively (Fitzgerald et al., 2007b). The level of methylation in the ATCV-1 genome is unknown. However, ATCV-1 only encodes one DNA methyltransferase (Z040R), which methylates cytosines. Therefore, the methylation of the ATCV-1 genome is expected to be low. The ATCV-1 DNA methyltransferase lacks a companion site-specific endonuclease.
Integration and transposition enzymes
Homing endonucleases are rare DNA-cleaving enzymes that are typically encoded by introns and inteins and the chlorella viruses encode many of these enzymes. Homing endonucleases are classified into four families (Belfort and Roberts, 1997). ATCV-1 encodes 17 homing endonucleases; 10 are members of the GIY-YIG family and 7 are members of the HNH family (Table 2). None of the chlorella virus-encoded homing endonucleases has been tested for activity. Therefore, it is unknown if they have an essential role in the replication cycle.
Unlike the NC64A viruses, ATCV-1 does not encode a transposase. However, like the other chlorella viruses, ATCV-1 encodes an ORF that resembles a Tlr 6Fp DNA mobile protein (Z016R). The Tlr 6Fp DNA mobile protein is encoded by a member of a family of genetic elements limited to Tetrahymena thermophila (Wuitschick et al., 2002).
Polyamine biosynthetic enzymes
PBCV-1 was the first virus reported to have genes encoding polyamine biosynthetic enzymes, including two pathways to synthesize putrescine. All four PBCV-1 genes, which encode functional enzymes (Kaiser et al., 1999; Morehead et al., 2002; Baumann et al., in press), are also present in ATCV-1. These enzymes are ornithine/arginine decarboxylase (ODC) (Z760R), agmatine iminohydrolase (Z806R), N-carbamoylputrescine amidohydrolase (Z169R), and homospermidine synthase (Z590L). ODC catalyzes the first and rate-limiting step in polyamine biosynthesis, the decarboxylation of ornithine to putrescine (Davis et al., 1992). The PBCV-1 ODC is the smallest ODC characterized to date (372 amino acids) (Morehead et al., 2002). Unexpectedly, the PBCV-1 enzyme decarboxylates arginine more efficiently than ornithine (Shah et al., 2004). ATCV-1 encodes a 372 amino acid ODC, with 59–72% amino acid identity to its chlorella virus homologs. The product of arginine decarboxylation is agmatine; agmatine iminohydrolase and N-carbamoylputrescine amidohydrolase convert agmatine to putrescine.
Homospermidine synthase synthesizes the rare polyamine homospermidine from two putrescine molecules (Kaiser et al., 1999). The ATCV-1 homospermidine synthase has 61–72% amino acid identity with its chlorella virus homologs.
ATCV-1 has genes encoding two additional enzymes involved in amine metabolism, monoamine oxidase (Z773L) and histidine decarboxylase (Z421L). The finding that ATCV-1 and each of the NC64A viruses have genes encoding these six proteins suggests that these enzymes must serve some important role(s) in virus replication.
Miscellaneous proteins
ATCV-1 has genes encoding several other putative proteins, including Cu/Zn superoxide dismutase (Z190L), amidase (Z177L), fibronectin-binding protein (Z119L), and an ABC transporter protein (Z086R). The Cu/Zn superoxide dismutase has 73–80% amino acid identity to the PBCV-1, MT325, and FR483-encoded homologs. This enzyme converts superoxide radicals into molecular oxygen and hydrogen peroxide (Bannister et al., 1987). Presumably, the ATCV-1 enzyme reduces light-induced superoxide accumulation. ATCV-1 has genes encoding two additional proteins that are unique to this virus, a mucin-desulfating sulfatase (Z734R) and an erythrocyte binding-like protein (Z092L).
ATCV-1 structural proteins
ATCV-1 ORF Z280L has the highest amino acid identity (78–81%) to and is approximately the same size as the PBCV-1 and the Pbi virus CVG-1 major capsid proteins (Plugge et al., 1999; Graves and Meints, 1992) Therefore, we assume that Z280L is the ATCV-1 major capsid protein. However, ATCV-1 encodes additional ORFs that have significant amino acid sequence identity to the PBCV-1 and CVG-1 major capsid proteins. For example, ATCV-1 ORFs Z151L, Z506L, Z558L, and Z664R have 29 to 43% amino acid identity to either the major capsid protein from PBCV-1 or from CVG-1.
Identification of gene families
Sixty-one of the ATCV-1 ORFs resemble one or more other ATCV-1 ORFs based on a blastp search with an E-value of less than 10−10, suggesting that they might be either gene families or gene duplications. This number is somewhat misleading, however, since some of these ORFs are grouped as families because they contain a common conserved domain, e.g., ankyrin repeats or a PAPK repeat, even though the remainder of the amino acid sequences differ. A total of 15 families have two members, 2 families have three members, 2 families have five members, 1 family has six members, and 1 family has nine members. One five-member family resembles the major capsid protein, the other five-member family has PAPK repeat domains, the six-member family resembles HNH endonucleases, and the nine-member family has GIY-YIG catalytic domains.
Conclusions
ATCV-1 is first virus to be sequenced that infects the endosymbiotic Chlorella SAG 3.83 isolated from the heliozoon A. turfacea (Bubeck and Pfitzner, 2005). The 288,047-bp ATCV-1 genome is predicted to encode 329 proteins as well as 11 tRNAs. The putative protein-encoding genes are relatively evenly distributed on both strands and intergenic space is minimal. Approximately 34% of the gene products have been identified; some resemble proteins from prokaryotes whereas others resemble eukaryotic proteins. Approximately 80% of the ATCV-1 gene products have homologs in the prototype chlorella virus, PBCV-1, suggesting that these proteins are important in virus replication. However, there are some interesting exceptions in which a gene is only present in ATCV-1, e.g., genes encoding dTDP-d-glucose 4,6 dehydratase, ribonucleoside-triphosphate reductase, and mucin-desulfating sulfatase.
Materials and methods
Viral DNA isolation and sequencing
Plaque-forming virus ATCV-1 was isolated from a fresh-water pond in Stuttgart, Germany in 2002. The ATCV-1 host, Chlorella SAG 3.83, was grown on MBBM medium (Bubeck and Pfitzner, 2005). The ATCV-1 virus was produced, purified, and the viral DNAs were isolated using methods and protocols developed for virus PBCV-1 (Bubeck and Pfitzner, 2005; Van Etten et al., 1981, 1983). The ATCV-1 genome was sequenced using a shot-gun strategy and dye-terminator chemistry on ABI3730xl automated DNA sequencers by Agencourt Biosciences (Beverly, MA). The genome was sequenced to 8-fold coverage of Phred 20 or greater bases. Final finishing and gap coverage were completed via primer walking from plasmid recombinant clones.
Genomic sequence analysis
A potential protein-encoding region, or ORF, was defined as a continuous stretch of DNA that translated into a polypeptide initiated by an ATG translation start codon and extended for 64 or more codons using the standard genetic code. The ORF Finder program (http://www.bioinformatics.org/sms/orf_find.html) was used to identify all potential ORFs that met this criterion. The ORFs were numbered consecutively starting at the beginning of the genome (as determined by alignment with the PBCV-1 genome). The letter R or L following the number indicates that the orientation of the putative ORF is either left-to-right or right-to-left, respectively.
Dot plots of the virus major ORFs were created to determine the orientation of the ATCV-1 genome relative to the PBCV-1 genome. Every major ORF was individually plotted against the PBCV-1 major ORFs using blastp (protein vs. protein). Similarities between the two ORFs with E-values <10−3 are presented. Putative tRNA genes were identified using the tRNAscan-SE program developed by Lowe and Eddy (1997). Gene families were identified when a major ORF had an E-value of less than 10−10 to another ORF within the same genome.
Analysis with public databases
Each identified ORF was used in a search for homologs using the protein–protein BLAST (blastp) program (Altschul et al., 1990) against the non-redundant (NR) protein databases at NCBI. The criterion used to search the NR database was as follows: Scoring matrix = blosum62. Each putative identified ORF was scanned for potential functional attributes using Pfam version 18.0 (Finn et al., 2006). Every identified ORF was additionally scanned to determine if it belonged to a particular COG. In each of the analyses, the top 10 results were recorded regardless of the E-values.
Nucleotide sequence accession number
The ATCV-1 sequence has been deposited in the GenBank database (accession number NC_008724 and the sequences can also be found at http://greengene.uml.edu.
Acknowledgments
We thank James Gurnon for preparing the DNA. This investigation was supported in part by National Science Foundation grant EF-0333197 (MG and JVE), by National Institutes of Health grant GM32441 (JVE), and by the Center of Biomedical Research Excellence program of the National Center for Research Resources Grant P20-RR15635 (JVE).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.virol.2006.12.028.
Footnotes
Publisher's Disclaimer: This article was originally published in a journal published by Elsevier, and the attached copy is provided by Elsevier for the author's benefit and for the benefit of the author's institution, for non-commercial research and educational use including without limitation use in instruction at your institution, sending it to specific colleagues that you know, and providing a copy to your institution's administrator.
References
- Ali AM, Kawasaki T, Yamada T. Genetic rearrangements on the chlorovirus genome that switch between hyaluronan synthesis and chitin synthesis. Virology. 2005;342:102–110. doi: 10.1016/j.virol.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bannister JV, Bannister WH, Rotilio G. Aspects of the structure, function, and applications of superoxide dismutase. CRC Rev. Biochem. 1987;22:111–180. doi: 10.3109/10409238709083738. [DOI] [PubMed] [Google Scholar]
- Baumann S, Sander A, Gurnon JR, Yanai-Balser G, Van Etten JL, Piotrowski M. Chlorella viruses contain genes encoding a complete polyamine biosynthetic pathway. Virology. doi: 10.1016/j.virol.2006.10.010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfield GP, Tuite MF. Translation elongation factor 3: a fungus-specific translation factor? Mol. Microbiol. 1993;9:411–418. doi: 10.1111/j.1365-2958.1993.tb01702.x. [DOI] [PubMed] [Google Scholar]
- Belfort M, Roberts RJ. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck JA, Pfitzner AJP. Isolation and characterization of a new type of chlorovirus that infects an endosymbiotic Chlorella strain of the heliozoon Acanthocystis turfacea. J. Gen. Virol. 2005;86:2871–2877. doi: 10.1099/vir.0.81068-0. [DOI] [PubMed] [Google Scholar]
- Chakraburtty K. Translational regulation by ABC systems. Res. Microbiol. 2001;152:391–399. doi: 10.1016/s0923-2508(01)01210-4. [DOI] [PubMed] [Google Scholar]
- Davis RH, Morris DR, Coffino P. Sequestered end products and enzyme regulation: the case of ornithine decarboxylase. Microbiol. Rev. 1992;56:280–290. doi: 10.1128/mr.56.2.280-290.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis PL, Wei J, Graves MV, Burbank DE, Van Etten JL. Hyaluronan synthase of chlorella virus PBCV-1. Science. 1997;278:1800–1803. doi: 10.1126/science.278.5344.1800. [DOI] [PubMed] [Google Scholar]
- Delaroque N, Muller DG, Bothe G, Pohl T, Knippers R, Boland W. The complete DNA sequence of the Ectocarpus siliculosus virus EsV-1 genome. Virology. 2001;287:112–132. doi: 10.1006/viro.2001.1028. [DOI] [PubMed] [Google Scholar]
- Dickey JS, Choi T-J, Van Etten JL, Osheroff N. Chlorella virus marburg topoisomerase II: high DNA cleavage activity as a characteristic of chlorella virus type II enzymes. Biochemistry. 2005;44:3899–3908. doi: 10.1021/bi047777f. [DOI] [PubMed] [Google Scholar]
- Dunigan DD, Fitzgerald LA, Van Etten JL. Phycodnaviruses: a peek at genetic diversity. Virus Res. 2006;117:119–132. doi: 10.1016/j.virusres.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald LA, Graves MV, Li X, Feldblyum T, Hartigan J, Van Etten JL. Sequence and annotation of the 314-kb MT325 and the 321-kb FR483 viruses that infect Chlorella Pbi. Virology. 2007a;358:459–471. doi: 10.1016/j.virol.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald LA, Graves MV, Li X, Feldblyum T, Nierman WC, Van Etten JL. The sequence and annotation of the 369-kb NY-2A and the 345-kb AR158 viruses that infect Chlorella NC64A. Virology. 2007b;358:472–484. doi: 10.1016/j.virol.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S, Kang M, Epimashko S, Van Etten JL, Dainty J, Thiel G, Moroni A. Chlorella virus MT325 encodes water and potassium channels that interact synergistically. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5355–5360. doi: 10.1073/pnas.0600848103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Keller W. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem. Sci. 2001;26:376–384. doi: 10.1016/s0968-0004(01)01827-8. [DOI] [PubMed] [Google Scholar]
- Grabherr R, Strasser P, Van Etten JL. The DNA polymerase gene from chlorella viruses PBCV-1 and NY-2A contains an intron with nuclear splicing sequences. Virology. 1992;188:721–731. doi: 10.1016/0042-6822(92)90527-v. [DOI] [PubMed] [Google Scholar]
- Graves MV, Meints RH. Characterization of the major capsid protein and cloning of its gene from algal virus PBCV-1. Virology. 1992;188:198–207. doi: 10.1016/0042-6822(92)90750-j. [DOI] [PubMed] [Google Scholar]
- Graves MV, Burbank DE, Roth R, Heuser J, DeAngelis PL, Van Etten JL. Hyaluronan synthesis in virus PBCV-1 infected chlorella-like green algae. Virology. 1999;257:15–23. doi: 10.1006/viro.1999.9628. [DOI] [PubMed] [Google Scholar]
- Graves MV, Bernadt CT, Cerny R, Van Etten JL. Molecular and genetic evidence for a virus-encoded glycosyltransferase involved in protein glycosylation. Virology. 2001;285:332–345. doi: 10.1006/viro.2001.0937. [DOI] [PubMed] [Google Scholar]
- Graziani S, Xia Y, Gurnon JR, Van Etten JL, Leduc D, Skouloubris S, Myllykallio H, Liebl U. Functional analysis of FAD-dependent thymidylate synthase ThyX from Paramecium bursaria chlorella virus-1. J. Biol. Chem. 2004;279:54340–54347. doi: 10.1074/jbc.M409121200. [DOI] [PubMed] [Google Scholar]
- Ho CK, Van Etten JL, Shuman S. Characterization of an ATP-dependent DNA ligase encoded by Chlorella virus PBCV-1. J. Virol. 1997;71:1931–1937. doi: 10.1128/jvi.71.3.1931-1937.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Kaiser A, Vollmert M, Tholl D, Graves MV, Gurnon JR, Xing W, Lisec AD, Nickerson KW, Van Etten JL. Chlorella virus PBCV-1 encodes a functional homospermidine synthase. Virology. 1999;263:254–262. doi: 10.1006/viro.1999.9972. [DOI] [PubMed] [Google Scholar]
- Kang M, Moroni A, Gazzarrini S, DiFrancesco D, Thiel G, Severino M, Van Etten JL. Small potassium ion channel proteins encoded by chlorella viruses. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5318–5324. doi: 10.1073/pnas.0307824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami H, Kawakami N. Behavior of a virus in a symbiotic system, Paramecium bursaria-zoochlorella. J. Protozool. 1978;25:217–225. [Google Scholar]
- Kawasaki T, Tanaka M, Fujie M, Usami S, Sakai K, Yamada T. Chitin synthesis in chlorovirus CVK2-infected chlorella cells. Virology. 2002;302:123–131. doi: 10.1006/viro.2002.1572. [DOI] [PubMed] [Google Scholar]
- Kim Y, Clark DJ. SWI/SNF-dependent long-range remodeling of yeast HIS3 chromatin. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15381–15386. doi: 10.1073/pnas.242536699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landstein D, Graves MV, Burbank DE, DeAngelis P, Van Etten JL. Chlorella virus PBCV-1 encodes functional glutamine:fructose-6-phosphate amidotransferase and UDP-glucose dehydrogenase enzymes. Virology. 1998;250:388–396. doi: 10.1006/viro.1998.9388. [DOI] [PubMed] [Google Scholar]
- Li Y, Lu Z, Sun L, Ropp S, Kutish GF, Rock DL, Van Etten JL. Analysis of 74 kb of DNA located at the right end of the 330-kb chlorella virus PBCV-1 genome. Virology. 1997;237:360–377. doi: 10.1006/viro.1997.8805. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzur KL, Farooq A, Zeng L, Plotnikova O, Koch AW, Zhou M-M. A dimeric viral SET domain methyltransferase specific to Lys27 of histone H3. Nat. Struct. Biol. 2003;10:187–196. doi: 10.1038/nsb898. [DOI] [PubMed] [Google Scholar]
- Meints RH, Van Etten JL, Kuczmarski D, Lee K, Ang B. Viral infection of the symbiotic chlorella-like alga present in Hydra viridis. Virology. 1981;113:698–703. doi: 10.1016/0042-6822(81)90198-7. [DOI] [PubMed] [Google Scholar]
- Morehead TA, Gurnon JR, Adams B, Nickerson KW, Fitzgerald LA, Van Etten JL. Ornithine decarboxylase encoded by chlorella virus PBCV-1. Virology. 2002;301:165–175. doi: 10.1006/viro.2002.1573. [DOI] [PubMed] [Google Scholar]
- Myllykallio H, Lipowski G, Leduc D, Filee J, Forterre P, Liebl U. An alternative flavin-dependent mechanism for thymidylate synthesis. Science. 2002;297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- Nishida K, Kawasaki T, Fujie M, Usami S, Yamada T. Aminoacylation of tRNAs encoded by chlorella virus CVK2. Virology. 1999;263:220–229. doi: 10.1006/viro.1999.9949. [DOI] [PubMed] [Google Scholar]
- Plugge B, Becker B, Wolf AH. Several genes in Chlorella virus strain CVG-1 encode putative virion components. J. Gen. Virol. 1999;80:1067–1072. doi: 10.1099/0022-1317-80-4-1067. [DOI] [PubMed] [Google Scholar]
- Plugge B, Gazzarrini S, Nelson M, Cerana R, Van Etten JL, Derst C, DiFrancesco D, Moroni A, Thiel G. A potassium channel protein encoded by chlorella virus PBCV-1. Science. 2000;287:1641–1644. doi: 10.1126/science.287.5458.1641. [DOI] [PubMed] [Google Scholar]
- Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Shah R, Coleman CS, Mir K, Baldwin J, Van Etten JL, Grishin NV, Pegg AE, Stanley BA, Phillips MA. Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of eukaryotic ornithine decarboxylases. J. Biol. Chem. 2004;279:35760–35767. doi: 10.1074/jbc.M405366200. [DOI] [PubMed] [Google Scholar]
- Sriskanda V, Shuman S. Role of nucleotidyl transferase motif V in strand joining by chlorella virus DNA ligase. J. Biol. Chem. 2002;277:9695–9700. doi: 10.1074/jbc.M110613200. [DOI] [PubMed] [Google Scholar]
- Sugimoto I, Onimatsu H, Fujie M, Usami S, Yamada T. vAL-1, a novel polysaccharide lyase encoded by chlorovirus CVK2. FEBS Lett. 2004;559:51–56. doi: 10.1016/S0014-5793(04)00022-5. [DOI] [PubMed] [Google Scholar]
- Sun L, Li Y, McCullough AK, Wood TG, Lloyd RS, Adams B, Gurnon JR, Van Etten JL. Intron conservation in a UV-specific DNA repair gene encoded by chlorella viruses. J. Mol. Evol. 2000;50:82–92. doi: 10.1007/s002399910009. [DOI] [PubMed] [Google Scholar]
- Tonetti M, Zanardi D, Gurnon JR, Fruscione F, Armirotti A, Damonte G, Sturla L, De Flora A, Van Etten JL. Paramecium bursaria chlorella virus 1 encodes two enzymes involved in the biosynthesis of GDP-l-fucose and GDP-d-rhamnose. J. Biol. Chem. 2003;278:21559–21565. doi: 10.1074/jbc.M301543200. [DOI] [PubMed] [Google Scholar]
- Van Etten JL, Meints RH, Burbank DE, Kuczmarski D, Cuppels DA, Lane LC. Isolation and characterization of a virus from the intracellular green alga symbiotic with Hydra viridis. Virology. 1981;113:704–711. doi: 10.1016/0042-6822(81)90199-9. [DOI] [PubMed] [Google Scholar]
- Van Etten JL, Meints RH, Kuczmarski D, Burbank DE, Lee K. Viruses of symbiotic chlorella-like algae isolated from Paramecium bursaria and Hydra viridis. Proc. Natl. Acad. Sci. U. S. A. 1982;79:3867–3871. doi: 10.1073/pnas.79.12.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten JL, Burbank DE, Xia Y, Meints RH. Growth cycle of a virus, PBCV-1, that infects chlorella-like algae. Virology. 1983;126:117–125. doi: 10.1016/0042-6822(83)90466-x. [DOI] [PubMed] [Google Scholar]
- Van Etten JL, Burbank DE, Joshi J, Meints RH. DNA synthesis in a chlorella-like alga following infection with the virus PBCV-1. Virology. 1984;134:443–449. doi: 10.1016/0042-6822(84)90311-8. [DOI] [PubMed] [Google Scholar]
- Van Etten JL, Schuster AM, Girton L, Burbank DE, Swinton D, Hattman S. DNA methylation of viruses infecting a eukaryotic chlorella-like alga. Nucleic Acids Res. 1985;13:3471–3478. doi: 10.1093/nar/13.10.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten JL, Lane LC, Meints RH. Viruses and viruslike particles of eukaryotic algae. Microbiol. Rev. 1991;55:586–620. doi: 10.1128/mr.55.4.586-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warbrick E. The puzzle of PCNA's many partners. BioEssays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Wilson WH, Schroeder DC, Allen MJ, Holden MT, Parkhill J, Barrell BG, Churcher C, Hamlin N, Mungall K, Norbertczak H, Quail MA, Price C, Rabbinowitsch E, Walker D, Craigon M, Roy D, Ghazal P. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005a;309:1090–1092. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- Wilson WH, Van Etten JL, Schroeder DS, Nagasaki K, Brussaard C, Delaroque N, Bratbak G, Suttle C. Phycodnaviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy: Classification and Nomenclature of Viruses. Eighth Report of the International Committee on the Taxonomy of Viruses. Elsevier; San Diego: 2005b. pp. 163–175. [Google Scholar]
- Wuitschick JD, Gershan JA, Lochowicz AJ, Li S, Karrer KM. A novel family of mobile genetic elements is limited to the germline genome in Tetrahymena thermophila. Nucleic Acids Res. 2002;30:2524–2537. doi: 10.1093/nar/30.11.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Fukuda T, Tamura K, Furukawa S, Songsri P. Expression of the gene encoding a translational elongation factor 3 homolog of chlorella virus CVK2. Virology. 1993;197:742–750. doi: 10.1006/viro.1993.1650. [DOI] [PubMed] [Google Scholar]
- Yamada T, Tamura K, Aimi T, Songsri P. Self-splicing group I introns in eukaryotic viruses. Nucleic Acids Res. 1994;22:2532–2537. doi: 10.1093/nar/22.13.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Onimatsu H, Van Etten JL. Chlorella viruses. In: Maramorosch K, Shatkin A, editors. Advances in Virus Research. Vol. 66. Elsevier; 2006. pp. 293–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Adams B, Sun L, Burbank DE, Van Etten JL. Intron conservation in the DNA polymerase gene encoded by chlorella viruses. Virology. 2001;285:313–321. doi: 10.1006/viro.2001.0935. [DOI] [PubMed] [Google Scholar]


