Abstract
The Rhizobium meliloti ExoK and ExsH glycanases have been proposed to contribute to production of low molecular weight (LMW) succinoglycan by depolymerizing high molecular weight succinoglycan chains in R. meliloti cultures. We expressed and purified ExoK and ExsH and determined that neither enzyme can extensively cleave succinoglycan prepared from R. meliloti cultures, although neutral/heat treatment and acid/heat treatment convert succinoglycan to forms that can be cleaved efficiently by both enzymes. These results were somewhat surprising, given that the exoK+ and exsH+ genes play a crucial role in production of LMW succinoglycan in R. meliloti cultures. We demonstrated by Western blot analyses that R. meliloti expresses ExoK and ExsH, that both proteins can be detected extracellularly, and that ExsH secretion depends on the prsD+/prsE+ genes, consistent with previous predictions based on mutant analyses. Furthermore, we determined that the depolymerization activities associated with purified ExoK and ExsH are comparable with exoK+ and exsH+-dependent depolymerization activities expressed in R. meliloti cultures. We resolved the apparent contradiction between the results of our previous genetic analyses and depolymerization assays by determining that ExoK and ExsH can cleave high molecular weight succinoglycan that is being produced actively by R. meliloti, but not succinoglycan that has accumulated in cultures, to yield LMW succinoglycan. We propose that ExoK and ExsH dynamically regulate the molecular weight distribution of succinoglycan by cleaving nascent succinoglycan only during a limited period after its synthesis, perhaps before it undergoes a time-dependent change in its conformation or aggregation state.
The nitrogen fixing symbioses established between soil bacteria of the Rhizobiaceae and leguminous plants are complex developmental processes involving the exchange of multiple signals between the bacteria and their hosts (1, 2). For rhizobial–legume symbioses that involve the formation of indeterminate root nodules, such as the R. meliloti–alfalfa symbiosis, bacterial production of extracellular polysaccharides (EPS) is crucial for successful invasion of root nodules (3–7). The precise roles played by bacterial EPS in the invasion process are not known, but evidence that particular forms of EPS exhibit specificity for particular symbioses (8, 9) and that small quantities of EPS are sufficient to mediate invasion by EPS deficient mutants (8, 10, 11) suggests the possibility that EPS plays a signaling role in invasion.
R. meliloti has the capacity to produce two types of EPS, succinoglycan (12, 13) and EPS II (9, 14–17) and a capsular polysaccharide, KPS (18). Each of these polysaccharides can be produced in either symbiotically active or symbiotically inactive forms (6, 9, 10, 18, 19). Of these polysaccharides, the wild-type strain Rm1021 produces only succinoglycan in a symbiotically active form and thus relies on succinoglycan production for invasion (6, 7). The structure of succinoglycan has been determined: Succinoglycan is a polymer of an octasaccharide-repeating unit, consisting of galactose, glucose, acetate, succinate, and pyruvate in a ratio of ≈1:7:1:1:1 (12, 13). In cultures, R. meliloti produces succinoglycan in high molecular weight (HMW) forms, consisting of hundreds to thousands of octasaccharide-repeating units, and low molecular weight (LMW) forms, consisting of monomers, trimers, and tetramers of the octasaccharide-repeating unit (11, 20, 21). LMW forms of succinoglycan have been proposed to be the forms crucial for establishment of symbiosis (11, 22).
The genetic analysis of succinoglycan production by R. meliloti has been greatly facilitated by the use of Calcofluor (Sigma), a fluorescent dye that binds succinoglycan. When colonies of the wild-type strain are cultivated on growth medium supplemented with Calcofluor and are visualized under UV light, the colonies fluoresce brightly because of Calcofluor binding to the succinoglycan that is produced by the colonies (6). Mutagenesis of the wild-type strain, followed by screening for nonfluorescing colonies, enabled the isolation of many mutants defective in production of succinoglycan (6, 23, 24). A group of genes involved in regulation, synthesis, and processing of succinoglycan have been cloned and sequenced, and most of these genes are located in a cluster termed the “exo” region (23–29). Based on analyses of the nucleotide sequence of the exo genes and characterization of the succinoglycan biosynthetic intermediates that accumulate in the cells of exo mutants, a model for succinoglycan biosynthesis has been proposed (30).
How R. meliloti controls the molecular weight distribution of succinoglycan is less well understood. Two simple models are that the bacteria produce LMW succinoglycan (i) by direct synthesis or (ii) by expressing glycanases that cleave HMW succinoglycan to yield LMW forms. These two models are not mutually exclusive; indeed evidence has been reported in support of both. Regarding the first model, truncation of the exoP gene, proposed to be involved in polymerization of succinoglycan, causes a marked decrease in the ratio of HMW:LMW succinoglycan produced by R. meliloti (31). Also, researchers have obtained evidence for genetically separable systems for the direct synthesis of LMW and HMW succinoglycan (J. E. González, C. E. Semino, L. E. Castellano-Torres, and G. C. Walker; unpublished results).
Regarding the second model, we have reported that R. meliloti strains with transposon mutations in the exoK gene and either the exsH, prsD, or prsE gene exhibit a dramatic defect in production of LMW succinoglycan (32). Based on nucleotide sequence analyses, ExoK and ExsH are predicted to belong to two distinct subclasses of endo-1,3–1,4-β-glycanases (26, 27, 32). Enzymes of this type would be predicted to cleave HMW succinoglycan to yield monomers of the octasaccharide-repeating unit, structurally identical to the octasaccharide subunits generated during succinoglycan biosynthesis (33), or multimers of the repeating unit. Thus, ExoK and ExsH are excellent candidates for proteins directly involved in depolymerization of HMW succinoglycan to yield LMW forms. Both ExoK and ExsH are predicted to accumulate extracellularly (32). In the case of ExsH in particular, its extracellular localization is predicted to be crucial for its activity, given that the prsD and prsE genes encode the ABC-type transporter and the membrane fusion protein of a type I secretion system, respectively, that the exsH gene encodes a domain typical of proteins secreted by type I secretion systems, and that mutations in any one of these three genes cause defects in production of LMW succinoglycan of the same magnitude (32).
To refine our model for how glycanases contribute to production of LMW succinoglycan, we tested ExoK and ExsH in reconstituted succinoglycan depolymerization reactions. We demonstrated that ExoK and ExsH can depolymerize succinoglycan prepared from R. meliloti cultures; although we found that treatments that alter the physical properties of succinoglycan greatly affect the extent to which ExoK and ExsH can cleave the polysaccharide. Strikingly, we determined also that ExoK and ExsH can cleave succinoglycan produced by actively growing cells, but not succinoglycan in cell-free culture supernatants, to yield LMW succinoglycan. We infer that the physiologically relevant role of these glycanases is to cleave nascent succinoglycan chains rather than to cleave succinoglycan, which has accumulated extracellularly.
MATERIALS AND METHODS
Strains, Growth Media, and Preparation of Succinoglycan.
The R. meliloti strains Rm1021 (wild-type), Rm7210 (exoY), Rm8832 (exoY exoK), Rm8833 (exoY exsH), Rm8834 (exoY exoK exsH), Rm8835 (exoY prsD), and Rm8836 (exoY prsE) (32) and the Escherichia coli strain BL21(DE3) (34) have been described. R. meliloti strains were cultivated at 30°C as described (11). The following growth media were used: Luria–Bertani (LB) (35), M9 (35), mannitol/glutamate/salts (MGS) (pH 7.4) (32), and glutamate/mannitol/salts (GMS) (32). Succinoglycan was prepared from MGS cultures of Rm1021 as described (36).
Expression, Recovery, and Analysis of ExoK and ExsH.
To construct pEXOK and pEXSH, we amplified the complete exoK and exsH ORFs by PCR while simultaneously introducing restriction sites (NdeI and BclI for exoK and NdeI and BamHI for exsH) at the ends of the fragments and then cloned these fragments into the vector pET5a (Promega), which had been digested with NdeI and BamHI. We used previously described techniques to induce expression of ExoK and ExsH in the strain BL21(DE3) (34). For preparation of ExoK, ExsH, and soluble protein from BL21(DE3)/pET5a, cells from 100 ml of culture were resuspended in buffer (50 mM KCl/50 mM Hepes, pH 7.5/1 mM EDTA/0.5% Nonidet P-40/1 mM DTT) and were lysed by sonication. ExoK and ExsH were isolated by denaturing insoluble cellular proteins in buffer (8 M urea/50 mM Hepes/50 mM DTT), dialyzing samples against buffer (5 M urea/25 mM Hepes/100 mM KCl) for 2 hr, and removing urea and DTT by further staged dialysis against buffer (25 mM Hepes/100 mM KCl). All of the protein preparation steps were conducted at 4°C. Protein samples were separated by discontinuous SDS/PAGE (10% polyacrylamide separating gels) and visualized by staining with Coomassie brilliant blue, as described (37).
Carbohydrate Analyses.
For in vitro depolymerization assays, succinoglycan (≈0.3 mg/ml) was dissolved in reaction buffer (50 mM potassium phosphate, pH 7.0/1 mM MgSO4/0.25 mM CaCl2), treated with ExoK, ExsH, or soluble protein from BL21(DE3)/pET5a (0.16–20 μg/ml final concentrations), and incubated at 30°C for 24 hr. Carbohydrate concentrations and relative reducing end concentrations were determined by the anthrone/sulfuric acid method (38) and the Lever method (39), respectively. We used previously described methods for Biogel A5-m and Biogel P4 column chromatography (Bio-Rad) (11, 20, 32).
To determine the reducing end generated by ExoK and ExsH-mediated cleavage of acid/heat-treated succinoglycan, enzyme-digested samples were passed over a Biogel P4 column and monomers of octasaccharide were recovered. Samples of 150 μg were then lyophilized, dissolved in 1 ml of water with 2 mg of sodium borohydride, incubated for 12 hr at room temperature, treated with 30 μl of 50% acetic acid, and desalted by passage over a Biogel P4 column. Reduced monomer samples were acid-hydrolyzed (2 M trifluoroacetic acid at 121°C for 2 hr), lyophilized to remove the acid, dissolved in water, and analyzed by HPLC–anion exchange chromatography coupled with pulsed amperometric detection (Dionex). To confirm that extracellular carbohydrate present in cultures was succinoglycan, we used essentially the same method as described above except that (i) extracellular carbohydrate was depolymerized completely as described (32) and (ii) the step of reduction by sodium borohydride was omitted.
RESULTS
Expression of ExoK and ExsH in E. coli.
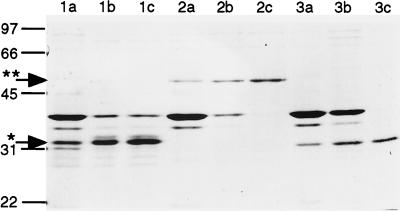
The R. meliloti exoK and exsH genes have been proposed to encode extracellular endo-1,3–1,4-β-glycanases of 30 and 50 kDa, respectively, that depolymerize HMW succinoglycan to yield LMW succinoglycan in R. meliloti culture supernatants (26, 27, 32). To reconstitute these depolymerization reactions in vitro, we used the pET expression system (Promega) to generate large quantities of ExoK and ExsH. Specifically, we amplified the complete exoK and exsH ORFs by PCR, cloned the amplified ORFs into the vector pET5a, confirmed that the two cloned ORFs were free of nucleotide sequence errors, and designated the two resulting plasmids pEXOK and pEXSH. We transferred each of the plasmids pEXOK, pEXSH, and pET5a into the E. coli strain BL21(DE3) and cultivated the resulting strains under conditions that induce expression of ORFs cloned into pET5a. We determined that the strain containing pEXOK expresses a pair of ≈31-kDa proteins that are unique to this strain (Fig. 1). These proteins correspond to full length or processed forms of ExoK. The strain containing pEXSH expresses a 51-kDa protein that corresponds to ExsH (Fig. 1). We have confirmed these assignments by Western blot analyses (see below). When expressed by the pET system, ExoK and ExsH are associated predominantly with proteins that remain insoluble after sonication of cells. We purified ExoK and ExsH to a substantial degree simply by subjecting these insoluble protein fractions to a denaturation/renaturation treatment followed by centrifugation to remove insoluble proteins. ExoK and ExsH comprise a high proportion of the remaining soluble proteins derived from the pEXOK and pEXSH containing strains, respectively.
Figure 1.
SDS/PAGE analysis and Coomasie blue staining of proteins expressed by BL21(DE3)/pEXOK (lanes 1a–c), BL21(DE3)/pEXSH (lanes 2a–c), or BL21(DE3)/pET5a (lanes 3a–c). Approximately 2 μg of protein was loaded per lane. Treatments: insoluble protein remaining after sonication of cells (lanes 1a, 2a, 3a), protein recovered after denaturation/renaturation procedure (lanes 1b, 2b, 3b) , and soluble fraction of renatured protein (lanes 1c, 2c, 3c). Lines indicate positions of molecular mass markers (kDa). Arrows indicate position of ExoK (∗) and ExsH (∗∗).
Native Succinoglycan Is Highly Refractory to Depolymerization by ExoK and ExsH.
We proceeded to test these soluble ExoK and ExsH preparations for succinoglycan depolymerase activity in vitro, under conditions relevant to the function of these enzymes in vivo. Reactions were incubated at 30°C, the standard growth temperature for R. meliloti. The substrate for these reactions was HMW succinoglycan purified from wild-type R. meliloti culture supernatants (native succinoglycan). And, because our previous analyses of R. meliloti strains implied that ExoK and ExsH function extracellularly, and that for ExsH in particular its extracellular localization is crucial for its activity, we used an assay buffer that simulates typical R. meliloti growth media.
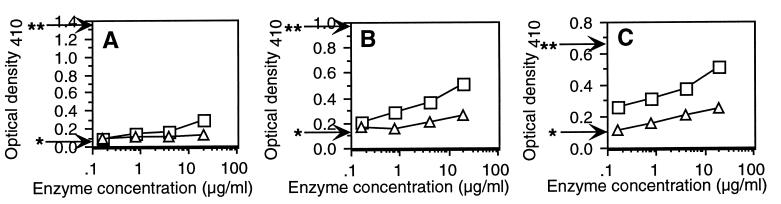
We made the striking observation that under these conditions ExoK and ExsH can cleave succinoglycan only to a slight extent. Treatment of native succinoglycan (375 μg/ml) with ExoK protein (20 μg/ml) for 24 hr yielded a 2.5-fold increase in the concentration of reducing ends, as determined by the Lever-reducing end assay (Fig. 2), and converted ≈3% of the HMW succinoglycan to LMW forms, as determined by Biogel P4 gel filtration chromatography. Similar treatment with ExsH protein (20 μg/ml) yielded a 1.2-fold increase in the concentration of reducing ends (Fig. 2) and also converted ≈3% of the HMW succinoglycan to LMW forms. Thus, neither ExoK nor ExsH were cleaving succinoglycan extensively. For comparison, succinoglycan that had been depolymerized completely to monomers of the octasaccharide-repeating unit by treatment with succinoglycan depolymerase from Cytophaga arvensicola (40) exhibited a 12-fold increase in the concentration of reducing ends (Fig. 2). It is worth noting that the Lever assay overestimates the concentration of reducing ends for polysaccharides, presumably because of alkaline hydrolysis of polysaccharide chains during the course of the assay (41); thus, we used the Lever assay here not to measure the precise degree of polymerization of samples but to provide a qualitative comparison of the extent of depolymerization among samples.
Figure 2.
Plot of OD410 vs. depolymerase concentration for samples of succinoglycan treated for 24 hr with either ExoK (squares) or ExsH (triangles) and then subjected to the Lever-reducing end assay. We tested a series of fivefold dilutions of the two proteins, starting at a concentration of 20 μg/ml. Substrates: 0.38 mg/ml native succinoglycan (A), 0.27 mg/ml neutral/heat-treated succinoglycan (B), and 0.20 mg/ml acid/heat-treated succinoglycan (C). Arrows indicate OD410 for untreated succinoglycan (*) or succinoglycan depolymerized completely to octasaccharide by succinoglycan depolymerase of Cytophaga arvensicola (∗∗). Each data point represents the average of three samples (SD < 10%).
We observed that ExoK and ExsH remained active over the 24-hr time course of the depolymerization reactions described above and that extending the reaction incubation times or supplementing reactions with additional ExoK or ExsH caused little or no increase in the extent of cleavage of succinoglycan. Thus, our results implied that a large proportion of the succinoglycan in these samples was refractory to cleavage by ExoK and ExsH. To gain insights into why the succinoglycan was refractory to cleavage, we heated succinoglycan solutions to 100°C for several minutes at neutral pH (neutral/heat treatment) or to 100°C for 90 min in 50 mM oxalic acid (acid/heat treatment), followed by cooling of the succinoglycan solutions to room temperature (and neutralization and dialysis of acid/heat-treated samples). These treatments irreversibly converted succinoglycan to forms that were more susceptible to cleavage by ExoK and ExsH (Fig. 2). For example, incubation of acid/heat-treated succinoglycan with ExoK (20 μg/ml) or ExsH (20 μg/ml) caused the conversion of 31 or 22% of the total succinoglycan to LMW forms, respectively. We determined that extracts of soluble proteins derived from the strain containing only the pET5a vector caused no cleavage of any form of succinoglycan. This control indicates that the cleavage of succinoglycan is due to ExoK or ExsH, respectively, and not due to any contaminating E. coli proteins present in the preparations. Furthermore, by purifying samples of depolymerized acid/heat-treated succinoglycan and subjecting them to sodium borohydride reduction, acid hydrolysis, and HPLC–anion exchange chromatography coupled with pulsed amperometric detection, we determined that both ExoK and ExsH cleave succinoglycan to yield galactose at the reducing end of the cleaved product (data not shown). Thus, we have demonstrated that ExoK and ExsH are endo-1,3–1,4-β-glycanases, as predicted by nucleotide sequence (26, 27, 32).
R. meliloti Expresses and Secretes ExoK and ExsH.
The observation that native succinoglycan is highly refractory to cleavage by ExoK and ExsH was somewhat surprising because our previous genetic analyses had implied that ExoK and ExsH cleave succinoglycan in culture supernatants (32). To help resolve this apparent contradiction, we raised polyclonal antibodies against ExoK and ExsH purified from the strains carrying pEXOK and pEXSH and used the antibodies to measure expression and secretion of ExoK and ExsH by R. meliloti. For these assays, we tested R. meliloti strains cultivated in two types of growth media, MGS, in which the exoK gene is the major determinant of Calcofluor halo production by R. meliloti colonies (23, 32), and GMS, in which the exsH/prsD/prsE genes are the major determinants of production of LMW succinoglycan by R. meliloti cells (32).
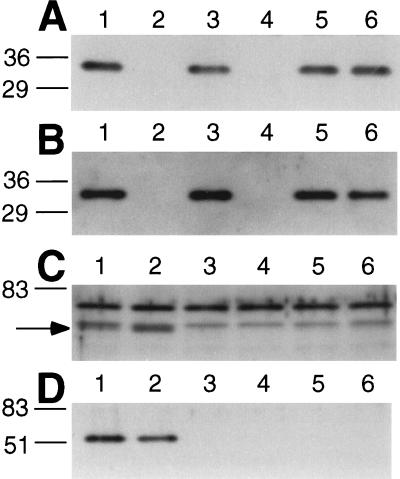
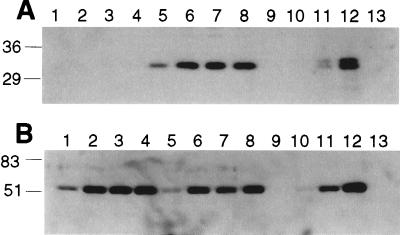
We determined that R. meliloti expresses ExoK. Antibodies against ExoK protein bind specifically to a 31-kDa protein (ExoK) that is expressed by R. meliloti cells with the exoK+ gene but that is not expressed by exoK mutants (Fig. 3). Although ExoK accumulates to approximately the same levels in cells cultivated in either MGS or GMS medium (data not shown), ExoK accumulates to ≈25-fold higher levels in culture supernatants in MGS medium (≈160 ng of ExoK per milliliter) than it does in GMS medium (≈6.4 ng of ExoK per milliliter) (Fig. 4).
Figure 3.
Western blot analyses of expression of ExoK (A and B) and ExsH (C and D) by R. meliloti strains. Lanes contain either the cells (without supernatant) from the equivalent of 10 μl of culture (A and C) or 5 μl of cell-free supernatant (B and D). Strains were cultured either in MGS medium (A and B) or GMS medium (C and D). We used exoY mutant strains, which fail to produce succinoglycan, to enable the high efficiency removal of cells from culture supernatants by centrifugation. Strains: 1, exoY; 2, exoY exoK; 3, exoY exsH; 4, exoY exoK exsH; 5, exoY prsD; and 6, exoY prsE. Lines indicate positions of molecular mass markers (kDa). Arrow for C indicates the position of protein band corresponding to ExsH, which is resolved poorly from another R. meliloti protein.
Figure 4.
Western blot analyses that compare the extent to which ExoK (A) and ExsH (B) accumulate in the culture supernatants of exoY strains cultivated in MGS vs. GMS medium. Lanes 1 through 8 contain 5 μl of cell-free culture supernatant from an exoY strain grown in GMS (lanes 1–4) or MGS (lanes 5–8). Supernatants were collected on day 1 (lanes 1 and 5), day 2 (lanes 2 and 6), day 3 (lanes 3 and 7), and day 5 (lanes 4 and 8) of incubation of cultures. We also loaded 5-μl aliquots of a fivefold dilution series of the soluble protein fraction of BL21(DE3)/pEXOK (A) or BL21(DE3)/pEXSH (B), corresponding to concentrations of 1.3 ng/ml (lane 9), 6.4 ng/ml (lane 10), 32 ng/ml (lane 11), and 160 ng/ml (lane 12). For both blots, we also loaded 5 μl of 160 ng/ml of soluble protein from BL21(DE3)/pET5a (lane 13) and confirmed that the antibodies used here bind specifically to ExoK and ExsH rather than to endogenous E. coli proteins.
We also determined that R. meliloti expresses ExsH. Antibodies raised against ExsH bind specifically to a 51-kDa protein (ExsH) that is present at high levels in culture supernatants of the wild-type strain but not in culture supernatants of exsH, prsD, or prsE mutants (Fig. 3). Whether strains are cultivated in MGS or GMS, ExsH accumulates in supernatants to approximately the same levels (≈160 ng ExsH/ml) (Fig. 4). ExsH also can be detected in R. meliloti cells of the wild-type strain but not in cells of exsH mutants (Fig. 3). The prsD and prsE mutants exhibit a substantial defect in the intracellular accumulation of ExsH, but in contrast to the case for exsH mutants, ExsH can be detected inside prsD and prsE mutants on prolonged exposure of Western blots to film (data not shown). Here, we have directly demonstrated that (i) R. meliloti expresses ExoK and ExsH, (ii) ExoK and ExsH accumulate in culture supernatants early during the growth phase of cultures, indicating that they are likely being secreted rather than diffusing from lysed cells, and (iii) secretion of ExsH but not ExoK is dependent on the prsD+/prsE+ genes (32). Our analyses also indicate that the concentrations of ExoK and ExsH in culture supernatants are at least 125-fold less than the highest concentrations of ExoK and ExsH we have tested in the in vitro depolymerization assays. Thus, we have ruled out the possibility that expression of extremely high levels of ExoK and ExsH by R. meliloti might account for the contribution of these enzymes to production of LMW succinoglycan in vivo.
By directly testing R. meliloti cultures for glycanase activity by use of a Congo Red dye (Sigma) assay, we determined that an exoK+-dependent glycanase activity can be detected for strains cultivated in MGS but not GMS medium and that an exsH+-dependent glycanase activity can be detected for strains cultivated in GMS but not MGS medium (unpublished results). The exoK result is consistent with our Western blot analyses (Fig. 4); ExoK is secreted to a much higher extent in MGS than in GMS cultures. The exsH result is consistent with the observation that purified ExsH is much more active in GMS than in MGS medium (data not shown). Our results indicate that, although growth conditions have a dramatic effect on the accumulation and activity of ExoK and ExsH in R. meliloti culture supernatants, the activities of ExoK and ExsH purified from E. coli are comparable with the activities of ExoK and ExsH expressed by R. meliloti.
ExoK and ExsH Efficiently Cleave Succinoglycan of Cells Actively Synthesizing Succinoglycan.
To explain the apparent contradiction between (i) the large contribution of the exoK+ gene and the exsH+/prsD+/prsE+ genes to production of LMW succinoglycan in R. meliloti cultures, and (ii) the low efficiency with which ExoK and ExsH cleave native succinoglycan, we reasoned that, in vivo, only nascent succinoglycan chains may be susceptible to cleavage and that they then become refractory to cleavage after a short time. To test this hypothesis, we treated a GMS medium culture of the R. meliloti exoK exsH strain with: (i) ExoK, (ii) ExsH, (iii) succinoglycan depolymerase of C. arvensicola, (iv) soluble proteins derived from the strain carrying pET5a, or (v) water, for a total of 24 hr. Treatments were conducted either in the presence of exoK exsH cells actively producing succinoglycan, by treating day 3 cultures or in the absence of exoK exsH cells, by removing cells from day 4 cultures and then treating the cell-free culture supernatants. Thus, for both sets of treatments, the cultures were incubating and producing succinoglycan for the same total amount of time.
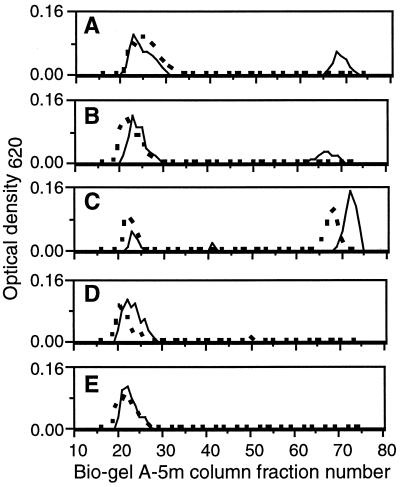
We observed that the addition to cultures of ExoK or ExsH protein caused 30 or 23%, respectively, of the total extracellular carbohydrate to accumulate as LMW forms (as determined by Biogel A5-m gel filtration chromatography) but only when the enzymes were added to cultures containing cells actively producing succinoglycan (Fig. 5). The addition of ExoK or ExsH to cell-free supernatants of day 3 exoK exsH cultures (data not shown) or cell-free supernatants of day 4 exoK exsH cultures caused no accumulation of LMW succinoglycan (Fig. 5). As expected, succinoglycan depolymerase from C. arvensicola caused the accumulation of LMW succinoglycan whether or not cells were present, and treatment with soluble proteins derived from the strain carrying pET5a or with water did not cause the accumulation of LMW succinoglycan in either case (Fig. 5). As a control, we confirmed that the LMW carbohydrate that accumulated in cultures treated with ExoK, ExsH, and succinoglycan depolymerase of C. arvensicola was actually succinoglycan rather than some other extracellular carbohydrate released by cells (data not shown). Our results directly demonstrate that ExoK and ExsH can contribute to production of LMW succinoglycan in R. meliloti cultures, presumably by depolymerizing HMW succinoglycan. Furthermore, the observation that cells must be present for efficient cleavage of succinoglycan by ExoK and ExsH in cultures, along with the results of our analyses of the activities of ExoK and ExsH produced by R. meliloti cultures, strongly suggests that ExoK and ExsH efficiently cleave succinoglycan chains only during a limited period after the newly synthesized succinoglycan chains emerge from the cell.
Figure 5.
Plot of OD620 vs. column fraction numbers, for cell-free culture supernatants of the exoK exsH strain cultivated in GMS, subjected to various treatments, and passed over a Biogel A5-m gel filtration column. Plots represent cultures from which cells were removed by centrifugation at day 4 before treatment (dashed line) or cultures that were treated at day 3 with cells still present (solid line). In both cases, treatments were conducted for 24 hr. In the latter case, cells were removed by centrifugation after the 24-hr treatment. Treatments: ExoK (20 μg/ml) (A), ExsH (20 μg/ml) (B), succinoglycan depolymerase of Cytophaga arvensicola (C), soluble protein from BL21(DE3)/pET5a (D), and water (E).
DISCUSSION
In the course of our experiments to test whether ExoK and ExsH are indeed succinoglycan depolymerases, we made an unexpected discovery. When added exogenously to cultures, both ExoK and ExsH are capable of efficiently cleaving succinoglycan from R. meliloti cells that actively are synthesizing the polysaccharide, but neither ExoK nor ExsH efficiently cleaves succinoglycan that is present in cell-free culture supernatants. The sum of our results leads to a striking inference; the physiologically relevant role of ExoK and ExsH in generating LMW succinoglycan is based on the enzymes specifically cleaving nascent succinoglycan rather than cleaving HMW succinoglycan that accumulates in the extracellular environment. Furthermore, ExoK and ExsH clearly are different from the succinoglycan depolymerase from C. arvensicola, which cleaves native succinoglycan extensively and which thus plays an important role in utilization of succinoglycan as a carbon source. The strikingly different properties of the R. meliloti glycanases suggest a different biological role.
The change that nascent succinoglycan undergoes that renders it insensitive to cleavage may be a time-dependent change in its conformation or aggregation state. Previous studies of the physical properties of succinoglycan may provide a useful framework for interpreting results from the depolymerization assays (42–45). Solutions of native succinoglycan are distinctive for their high viscosity, which may be attributed to nonuniform helicity of chains and/or lateral aggregation of chains (particularly at high succinoglycan concentrations) (42–45). Neutral/heat treatment of succinoglycan causes the succinoglycan to undergo a conformational transition resulting in an irreversible decrease in the viscosity of solutions upon cooling (42–44); this decrease correlates with conversion of succinoglycan to aggregates of uniformly helical chains (45). Acid/heat treatment of succinoglycan causes an even greater decrease in viscosity, which has been attributed to shortening of chains (due to acid hydrolysis) as well as to a decreased tendency of chains to aggregate (42). Because our depolymerization results clearly establish that treatments that cause a decrease in viscosity of succinoglycan solutions correlate with an increase in susceptibility of succinoglycan to cleavage by ExoK and ExsH, we speculate that the extent of helicity and/or aggregation of succinoglycan chains determine the extent to which succinoglycan can be cleaved by ExoK and ExsH.
The physical properties of succinoglycan also may have important biological consequences regarding the production of LMW succinoglycan in R. meliloti cultures. In the absence of salt, aggregation of succinoglycan occurs only at high succinoglycan concentrations (>1 mg/ml), but in the presence of even low concentrations of salt (10 mM sodium chloride), aggregation can occur at much lower succinoglycan concentrations (45). Based on the concentrations of salts in MGS and GMS media, succinoglycan would be predicted to aggregate in R. meliloti culture supernatants. One possible explanation for the high efficiency with which ExoK and ExsH cleave succinoglycan of cells actively synthesizing the polysaccharide is that nascent succinoglycan chains are highly susceptible to cleavage by ExoK and ExsH before aggregation of the chains. An endochitinase has been demonstrated to exhibit at least 80-fold greater activity and to yield a different set of cleaved oligosaccharide products when cleaving nascent chitin chains vs. chitin chains present in insoluble aggregates (46). However, the situation must be different with respect to succinoglycan; because all forms of succinoglycan are soluble, the change in the conformation or state of aggregation must be more subtle.
Our results indicate that the type of growth medium used can exert dramatic posttranslational effects on the accumulation and activity of the R. meliloti glycanases. ExoK, which may be secreted by the type II secretion pathway (32), accumulates to ≈25-fold greater amounts in R. meliloti culture supernatants in MGS vs. GMS medium. This observation raises the possibility that R. meliloti may regulate the secretion of certain proteins, such as ExoK, in response to environmental conditions. Such an ability to regulate the nature of proteins it secretes could be an important factor underlying the various bacterial–plant interactions that occur during the development of a productive symbiosis. ExsH, which is secreted by a type I secretion system (32), accumulates to approximately the same level in either medium, but the enzyme is much more active in GMS vs. MGS. The decreased activity of ExsH in MGS may be caused by the effects of growth medium components on the enzyme itself or on the physical properties of the substrate, succinoglycan.
Why does R. meliloti produce a polysaccharide and also express glycanases that depolymerize it? Perhaps this is a reflection of the importance of LMW succinoglycan in establishment of symbiosis. Although mutant analyses indicate that the exoK, exsH, prsD, and prsE genes are not required for establishment of symbiosis in an otherwise wild-type background (23, 32), it is possible that in the absence of ExoK and ExsH other mechanisms, such as direct synthesis of LMW succinoglycan, provide sufficient LMW succinoglycan for establishment of symbiosis. Alternatively, the production of a polysaccharide and a corresponding glycanase may provide a more general selective advantage to bacteria. Several other strains of bacteria, such as R. leguminosarum bv. viciae (47), Bradyrhizobium japonicum (48), and Pseudomonas marginalis (49), have been found to exhibit this trait, indicating that the phenomenon may be widespread.
Acknowledgments
We thank Phillips W. Robbins and Brett Pellock for critical reading of the manuscript. We thank M. A. Glucksmann for conducting preliminary analyses on ExoK glycanase activity. This work was supported by Public Health Service Grant GM31030 from the National Institutes of Health.
ABBREVIATIONS
- LMW
low molecular weight
- HMW
high molecular weight
- EPS
extracellular polysaccharides
- MGS
mannitol/glutamate/salts
- GMS
glutamate/mannitol/salts
References
- 1.Fisher R F, Long S R. Nature (London) 1992;357:655–660. doi: 10.1038/357655a0. [DOI] [PubMed] [Google Scholar]
- 2.Long S R. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borthakur D, Barber C E, Lamb J W, Daniels M J, Downie J A, Johnston A W B. Mol Gen Genet. 1986;203:320–323. [Google Scholar]
- 4.Chakravorty A K, Zurkowski W, Shine J, Rolfe B G. J Mol Appl Genet. 1982;1:585–596. [PubMed] [Google Scholar]
- 5.Chen H, Batley M, Redmond J W, Rolfe B G. J Plant Physiol. 1985;120:331–349. [Google Scholar]
- 6.Leigh J A, Signer E R, Walker G C. Proc Natl Acad Sci USA. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finan T M, Hirsch A M, Leigh J A, Johansen E, Kuldau G A, Deegan S, Walker G C, Signer E R. Cell. 1985;40:869–877. doi: 10.1016/0092-8674(85)90346-0. [DOI] [PubMed] [Google Scholar]
- 8.Djordjevic S P, Chen H, Batley M, Redmond J W, Rolfe B G. J Bacteriol. 1987;169:53–60. doi: 10.1128/jb.169.1.53-60.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glazebrook J, Walker G C. Cell. 1989;56:661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- 10.González J E, Reuhs B L, Walker G C. Proc Natl Acad Sci USA. 1996;93:8636–8641. doi: 10.1073/pnas.93.16.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Battisti L, Lara J C, Leigh J A. Proc Natl Acad Sci USA. 1992;89:5625–5629. doi: 10.1073/pnas.89.12.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansson P-E, Kenne L, Lindberg B, Ljunggren H, Ruden U, Svensson S. J Am Chem Soc. 1977;99:3812–3815. doi: 10.1021/ja00453a049. [DOI] [PubMed] [Google Scholar]
- 13.Aman P, McNeil M, Franzen L-E, Darvill A G, Albersheim P. Carbohydr Res. 1981;95:263–282. [Google Scholar]
- 14.Reinhold B B, Chan S Y, Reuber T L, Marra A, Walker G C, Reinhold V N. J Bacteriol. 1994;176:1997–2002. doi: 10.1128/jb.176.7.1997-2002.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Her G-R, Glazebrook J, Walker G C, Reinhold V N. Carbohydr Res. 1990;198:305–312. doi: 10.1016/0008-6215(90)84300-j. [DOI] [PubMed] [Google Scholar]
- 16.Keller M, Arnold W, Kapp D, Müller P, Niehaus K, Shmidt M, Quandt J, Weng W M, Pühler A. In: Rhizobium meliloti Genes Involved in Exopolysaccharide Production and Infection of Alfalfa Nodules. Silver S, Chakrabarty A M, Iglewski B, Kaplan S, editors. Washington, DC: American Society Microbiology; 1990. pp. 91–97. [Google Scholar]
- 17.Zhan H, Levery S B, Lee C C, Leigh J A. Proc Natl Acad Sci USA. 1989;86:3055–3059. doi: 10.1073/pnas.86.9.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reuhs B L, Carlson R W, Kim J S. J Bacteriol. 1993;175:3570–3580. doi: 10.1128/jb.175.11.3570-3580.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leigh J A, Reed J W, Hanks J F, Hirsch A M, Walker G C. Cell. 1987;51:579–587. doi: 10.1016/0092-8674(87)90127-9. [DOI] [PubMed] [Google Scholar]
- 20.Leigh J A, Lee C C. J Bacteriol. 1988;170:3327–3332. doi: 10.1128/jb.170.8.3327-3332.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breedveld M W, Zevenhuizen L P T M, Zehnder A J B. J Gen Microbiol. 1990;136:2511–2519. [Google Scholar]
- 22.Urzainqui A, Walker G C. J Bacteriol. 1992;174:3403–3406. doi: 10.1128/jb.174.10.3403-3406.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long S, Reed J W, Himawan J, Walker G C. J Bacteriol. 1988;170:4239–4248. doi: 10.1128/jb.170.9.4239-4248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller P, Hynes M, Kapp D, Niehaus K, Pühler A. Mol Gen Genet. 1988;211:17–26. [Google Scholar]
- 25.Glucksmann M A, Reuber T L, Walker G C. J Bacteriol. 1993;175:7033–7044. doi: 10.1128/jb.175.21.7033-7044.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glucksmann M A, Reuber T L, Walker G C. J Bacteriol. 1993;175:7045–7055. doi: 10.1128/jb.175.21.7045-7055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker A, Kleickmann A, Arnold W, Pühler A. Molec Gen Genet. 1993;238:145–154. doi: 10.1007/BF00279541. [DOI] [PubMed] [Google Scholar]
- 28.Becker A, Kleickmann A, Keller M, Arnold W, Pühler A. Mol Gen Genet. 1993;241:367–379. doi: 10.1007/BF00284690. [DOI] [PubMed] [Google Scholar]
- 29.Becker A, Kleickmann A, Küster H, Keller M, Arnold W, Pühler A. Mol Plant-Microbe Interact. 1993;6:735–744. doi: 10.1094/mpmi-6-735. [DOI] [PubMed] [Google Scholar]
- 30.Reuber T L, Walker G C. Cell. 1993;74:269–280. doi: 10.1016/0092-8674(93)90418-p. [DOI] [PubMed] [Google Scholar]
- 31.Becker A, Niehaus K, Pühler A. Mol Microbiol. 1995;16(2):191–203. doi: 10.1111/j.1365-2958.1995.tb02292.x. [DOI] [PubMed] [Google Scholar]
- 32.York G M, Walker G C. Mol Microbiol. 1997;25:117–134. doi: 10.1046/j.1365-2958.1997.4481804.x. [DOI] [PubMed] [Google Scholar]
- 33.Tolmasky M E, Staneloni R J, Leloir L F. J Biol Chem. 1982;257:6751–6757. [PubMed] [Google Scholar]
- 34.Studier F W, Moffatt B A. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 35.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 36.Ridout M J, Brownsey G J, York G M, Walker G C, Morris V J. Int J Biol Macromol. 1997;20:1–7. doi: 10.1016/s0141-8130(96)01140-3. [DOI] [PubMed] [Google Scholar]
- 37.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 38.Loewus F A. Anal Chem. 1952;24:219. [Google Scholar]
- 39.Lever M. Anal Biochem. 1972;47:273–279. doi: 10.1016/0003-2697(72)90301-6. [DOI] [PubMed] [Google Scholar]
- 40.Harada T. In: Determination of the Structure of β-d-Glycans from Strains of Agrobacterium and Rhizobium. BeMiller J N, Manners D J, Sturgeon R J, editors. X. New York: Wiley; 1994. pp. 155–163. [Google Scholar]
- 41.Greenwood C T, Milne E A. In: Starch Degrading and Synthesizing Enzymes: A Discussion of Their Properties and Action Pattern. Wolfrom M L, Tipson R S, editors. Vol. 23. New York: Academic; 1968. pp. 282–366. [DOI] [PubMed] [Google Scholar]
- 42.Fidanza M, Dentini M, Crescenzi V, Del Vecchio P. Int J Biol Macromol. 1989;11:372–376. doi: 10.1016/0141-8130(89)90010-x. [DOI] [PubMed] [Google Scholar]
- 43.Gravanis G, Milas M, Rinaudo M, Clarke-Sturman A J. Int J Biol Macromol. 1990;12:195–200. doi: 10.1016/0141-8130(90)90032-6. [DOI] [PubMed] [Google Scholar]
- 44.Gravanis G, Milas M, Rinaudo M, Clarke-Sturman A J. Int J Biol Macromol. 1990;12:201–206. doi: 10.1016/0141-8130(90)90033-7. [DOI] [PubMed] [Google Scholar]
- 45.Burova T V, Golubeva I A, Grinberg N V, Mashkevich A Y, Grinberg V Y, Usov A I, Navarini L, Cesáro A. Biopolymers. 1996;39:517–529. [Google Scholar]
- 46.Molano J, Polacheck I, Duran A, Cabib E. J Biol Chem. 1979;254:4901–4907. [PubMed] [Google Scholar]
- 47.Finnie C, Hartley N M, Findlay K C, Downie J A. Mol Microbiol. 1997;25:135–146. doi: 10.1046/j.1365-2958.1997.4471803.x. [DOI] [PubMed] [Google Scholar]
- 48.Dunn M F, Karr A L. Curr Microbiol. 1990;20:359–363. [Google Scholar]
- 49.Osman S F, Fett W F, Irwin P L, Bailey D G, Parris N, O’Connor J V. Curr Microbiol. 1993;26:299–304. [Google Scholar]