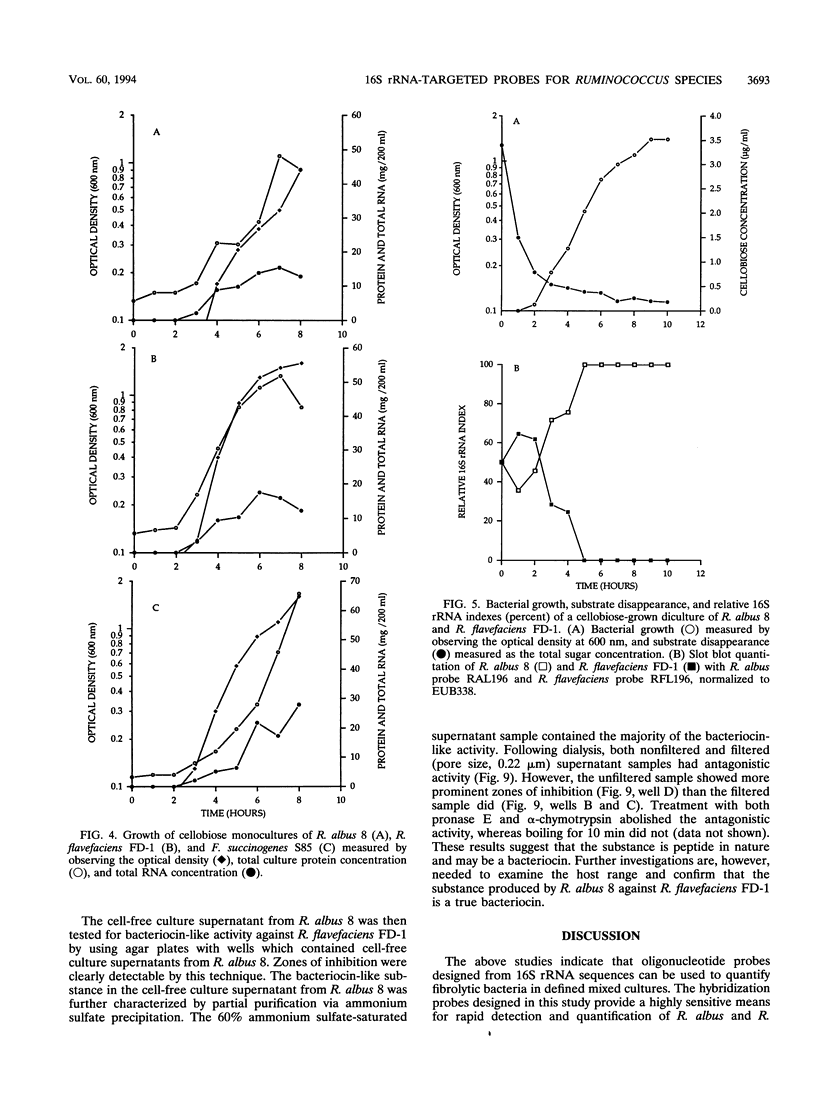
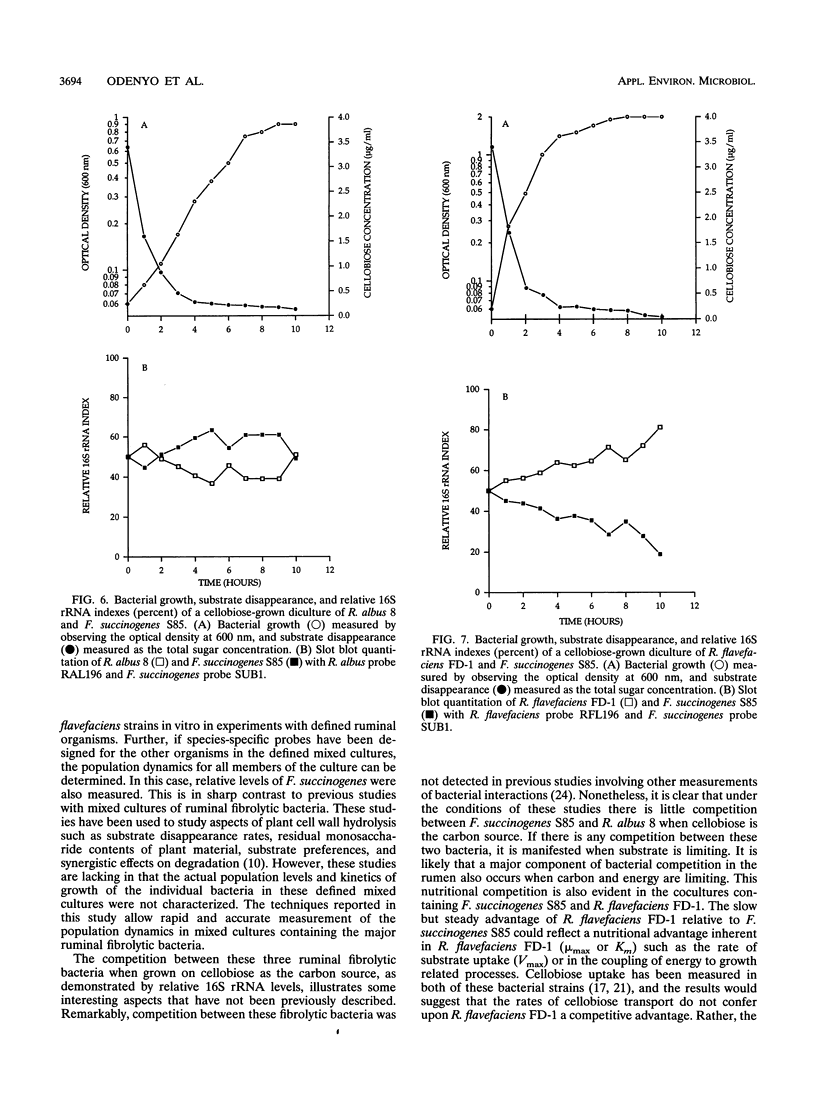
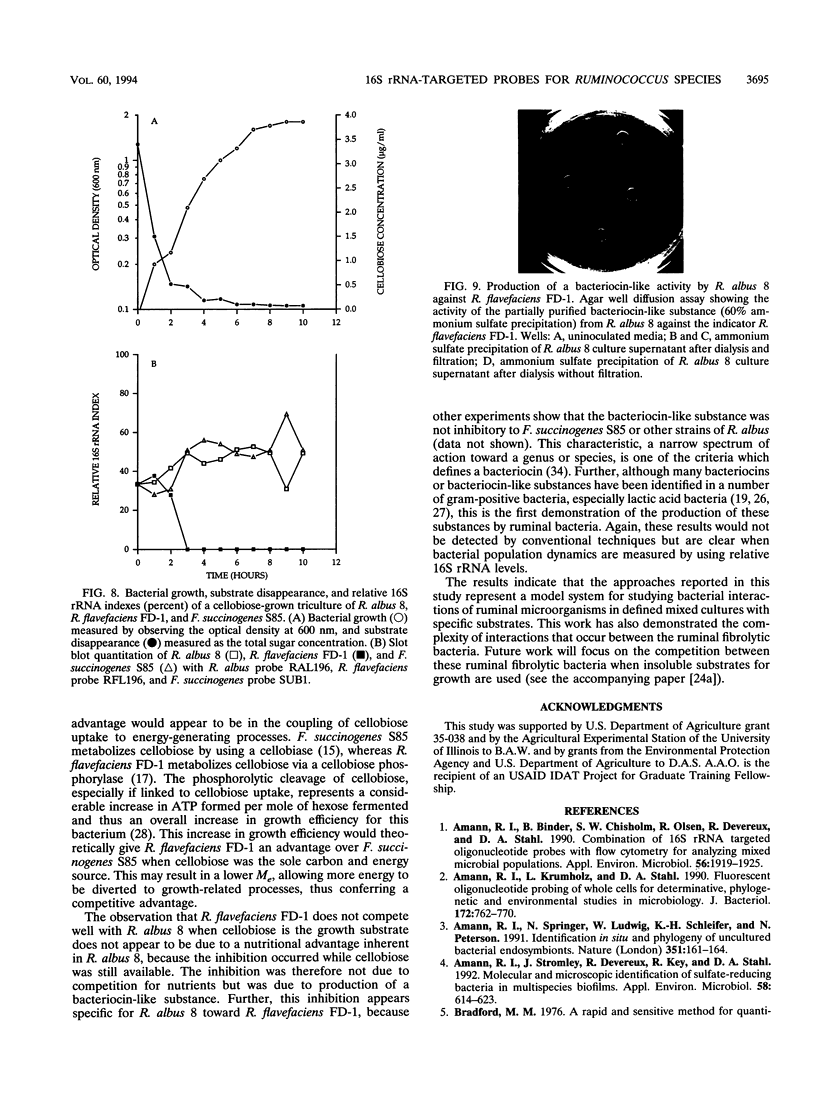
Abstract
A total of six oligonucleotide probes, complementary to the 16S rRNA, were evaluated for quantitative and determinative studies of Ruminococcus albus and Ruminococcus flavefaciens. On the basis of specificity studies, probes for R. albus (probe RAL196) and R. flavefaciens (probe RFL196) were selected to quantitate these species in mixed culture. In combination with a Fibrobacter succinogenes S85 subspecies probe (SUB1) and a domain Bacteria (formerly kingdom Eubacteria) probe (EUB338), they were used to quantitate these species competing in mixed cultures for cellobiose as the carbon source. In dicultures containing R. albus 8 and F. succinogenes S85, competition was not observed. However, R. flavefaciens FD-1 eventually outcompeted F. succinogenes S85 when cellobiose was the substrate. When R. albus 8 and R. flavefaciens FD-1 were grown together on cellobiose medium, R. albus 8 outcompeted R. flavefaciens FD-1, resulting in undetectable R. flavefaciens 16S rRNA only 1 to 3 h after inoculation, suggesting production of an antagonistic compound by R. albus 8 during rapid growth on soluble substrates. Further, when R. albus 8, R. flavefaciens FD-1, and F. succinogenes S85 were grown together in a triculture, R. flavefaciens FD-1 16S rRNA was detectable for only 2 h after inoculation, while R. albus 8 and F. succinogenes S85 showed a similar competition pattern to that of the dicultures. The results show that the Ruminococcus probes were effective in the measurement of relative populations of selected R. albus and R. flavefaciens strains during in vitro competition studies with F. succinogenes.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990 Jun;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Krumholz L., Stahl D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990 Feb;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Stromley J., Devereux R., Key R., Stahl D. A. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992 Feb;58(2):614–623. doi: 10.1128/aem.58.2.614-623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R., Springer N., Ludwig W., Görtz H. D., Schleifer K. H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991 May 9;351(6322):161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- Daba H., Pandian S., Gosselin J. F., Simard R. E., Huang J., Lacroix C. Detection and activity of a bacteriocin produced by Leuconostoc mesenteroides. Appl Environ Microbiol. 1991 Dec;57(12):3450–3455. doi: 10.1128/aem.57.12.3450-3455.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey G. P., Richardson B. C. Purification and Some Properties of Diplococcin from Streptococcus cremoris 346. Appl Environ Microbiol. 1981 Jan;41(1):84–89. doi: 10.1128/aem.41.1.84-89.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E. F., Wickham G. S., Pace N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989 Mar 10;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- Geis A., Singh J., Teuber M. Potential of lactic streptococci to produce bacteriocin. Appl Environ Microbiol. 1983 Jan;45(1):205–211. doi: 10.1128/aem.45.1.205-211.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groleau D., Forsberg C. W. Cellulolytic activity of the rumen bacterium Bacteroides succinogenes. Can J Microbiol. 1981 May;27(5):517–530. doi: 10.1139/m81-077. [DOI] [PubMed] [Google Scholar]
- Hahn D., Starrenburg M. J., Akkermans A. D. Oligonucleotide Probes That Hybridize with rRNA as a Tool To Study Frankia Strains in Root Nodules. Appl Environ Microbiol. 1990 May;56(5):1342–1346. doi: 10.1128/aem.56.5.1342-1346.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helaszek C. T., White B. A. Cellobiose uptake and metabolism by Ruminococcus flavefaciens. Appl Environ Microbiol. 1991 Jan;57(1):64–68. doi: 10.1128/aem.57.1.64-68.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King W., Raposa S., Warshaw J., Johnson A., Halbert D., Klinger J. D. A new colorimetric nucleic acid hybridization assay for Listeria in foods. Int J Food Microbiol. 1989 Jun;8(3):225–232. doi: 10.1016/0168-1605(89)90017-2. [DOI] [PubMed] [Google Scholar]
- Klaenhammer T. R. Bacteriocins of lactic acid bacteria. Biochimie. 1988 Mar;70(3):337–349. doi: 10.1016/0300-9084(88)90206-4. [DOI] [PubMed] [Google Scholar]
- Lathe R. Synthetic oligonucleotide probes deduced from amino acid sequence data. Theoretical and practical considerations. J Mol Biol. 1985 May 5;183(1):1–12. doi: 10.1016/0022-2836(85)90276-1. [DOI] [PubMed] [Google Scholar]
- Maas L. K., Glass T. L. Cellobiose uptake by the cellulolytic ruminal anaerobe Fibrobacter (Bacteroides) succinogenes. Can J Microbiol. 1991 Feb;37(2):141–147. doi: 10.1139/m91-021. [DOI] [PubMed] [Google Scholar]
- McSweeney C. S., Mackie R. I., Odenyo A. A., Stahl D. A. Development of an oligonucleotide probe targeting 16S rRNA and its application for detection and quantitation of the ruminal bacterium Synergistes jonesii in a mixed-population chemostat. Appl Environ Microbiol. 1993 May;59(5):1607–1612. doi: 10.1128/aem.59.5.1607-1612.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenyo A. A., Mackie R. I., Fahey G. C., Jr, White B. A. Degradation of wheat straw and alkaline hydrogen peroxide-treated wheat straw by Ruminococcus albus 8 and Ruminococcus flavefaciens FD-1. J Anim Sci. 1991 Feb;69(2):819–826. doi: 10.2527/1991.692819x. [DOI] [PubMed] [Google Scholar]
- Odenyo A. A., Mackie R. I., Stahl D. A., White B. A. The use of 16S rRNA-targeted oligonucleotide probes to study competition between ruminal fibrolytic bacteria: pure-culture studies with cellulose and alkaline peroxide-treated wheat straw. Appl Environ Microbiol. 1994 Oct;60(10):3697–3703. doi: 10.1128/aem.60.10.3697-3703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Overbeek R., Larsen N., Marsh T. L., McCaughey M. J., Maciukenas M. A., Kuan W. M., Macke T. J., Xing Y., Woese C. R. The Ribosomal Database Project. Nucleic Acids Res. 1992 May 11;20 (Suppl):2199–2200. doi: 10.1093/nar/20.suppl.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piard J. C., Muriana P. M., Desmazeaud M. J., Klaenhammer T. R. Purification and Partial Characterization of Lacticin 481, a Lanthionine-Containing Bacteriocin Produced by Lactococcus lactis subsp. lactis CNRZ 481. Appl Environ Microbiol. 1992 Jan;58(1):279–284. doi: 10.1128/aem.58.1.279-284.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M. A., Hespell R. B., White B. A., Bothast R. J. Inhibitory Effects of Methylcellulose on Cellulose Degradation by Ruminococcus flavefaciens. Appl Environ Microbiol. 1988 Apr;54(4):890–897. doi: 10.1128/aem.54.4.890-897.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Flesher B., Mansfield H. R., Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988 May;54(5):1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Lane D. J., Olsen G. J., Pace N. R. Analysis of hydrothermal vent-associated symbionts by ribosomal RNA sequences. Science. 1984 Apr 27;224(4647):409–411. doi: 10.1126/science.224.4647.409. [DOI] [PubMed] [Google Scholar]
- Stahl D. A., Lane D. J., Olsen G. J., Pace N. R. Characterization of a Yellowstone hot spring microbial community by 5S rRNA sequences. Appl Environ Microbiol. 1985 Jun;49(6):1379–1384. doi: 10.1128/aem.49.6.1379-1384.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagg J. R., Dajani A. S., Wannamaker L. W. Bacteriocins of gram-positive bacteria. Bacteriol Rev. 1976 Sep;40(3):722–756. doi: 10.1128/br.40.3.722-756.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien H. C., Bratina B. J., Tsuji K., Hanson R. S. Use of oligodeoxynucleotide signature probes for identification of physiological groups of methylotrophic bacteria. Appl Environ Microbiol. 1990 Sep;56(9):2858–2865. doi: 10.1128/aem.56.9.2858-2865.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]