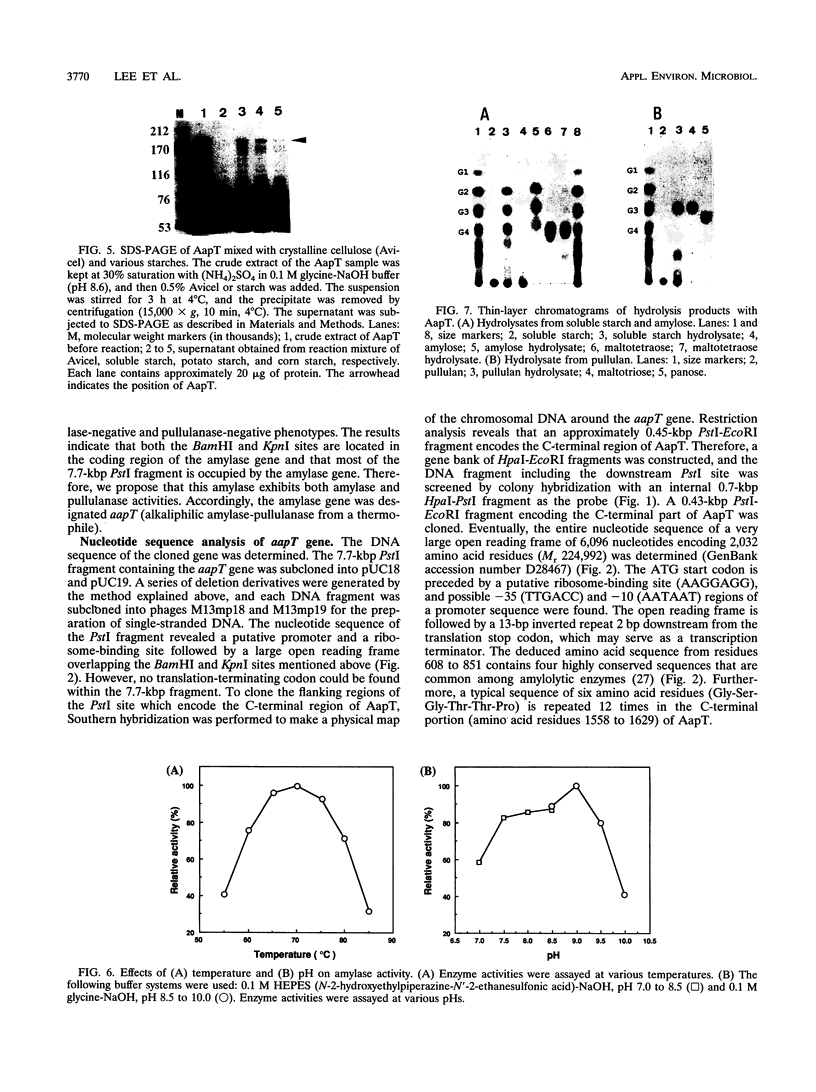
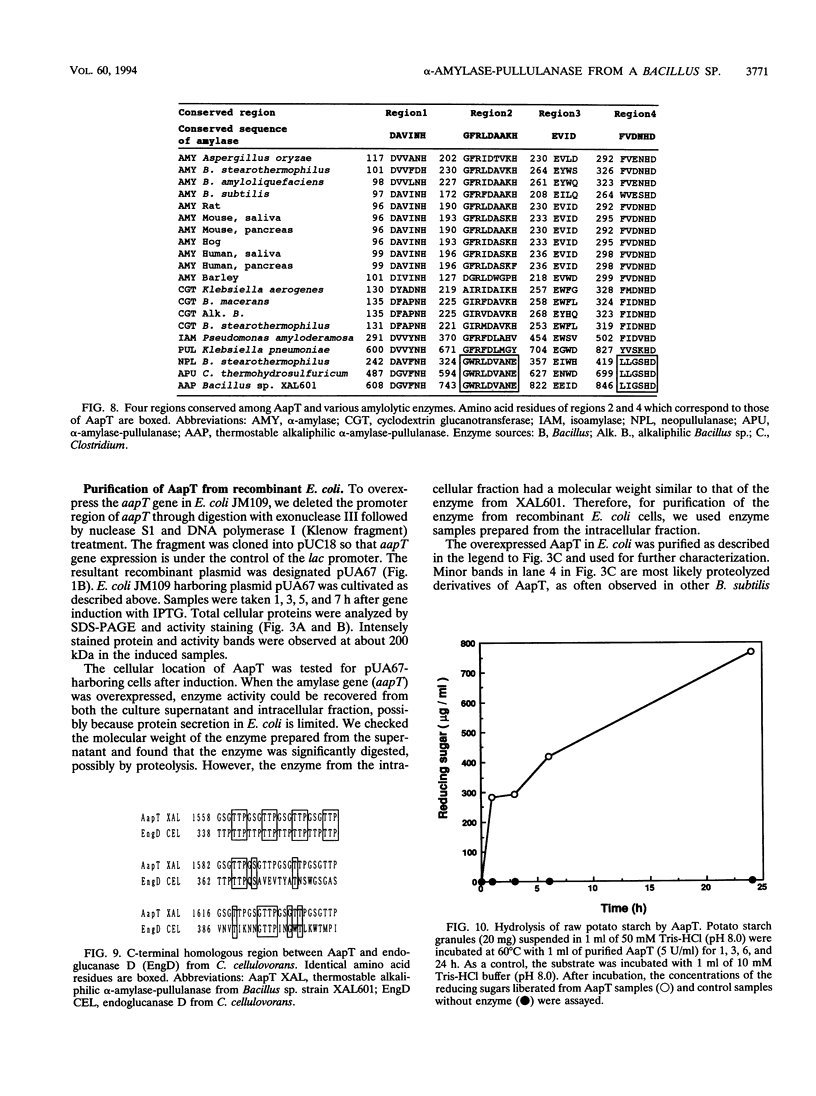
Abstract
A thermophilic and alkaliphilic Bacillus sp. strain, XAL601, was isolated from soil. It produces a thermostable and alkaline-stable enzyme with both alpha-amylase and pullulanase activities. The alpha-amylase-pullulanase gene (aapT) from this Bacillus strain was cloned, and its nucleotide sequence was determined (GenBank accession number D28467). A very large open reading frame composed of 6,096 bases, which encodes 2,032 amino acid residues with an M(r) of 224,992, was found. The deduced amino acid sequence revealed that the four highly conserved regions that are common among amylolytic enzymes were well conserved. These include an active center and common substrate-binding sites of various amylases. In the C-terminal region, a six-amino-acid sequence (Gly-Ser-Gly-Thr-Thr-Pro) is repeated 12 times. The aapT gene was then subcloned in Escherichia coli and overexpressed under the control of the lac promoter. Purification of AapT from this recombinant E. coli was performed, and it was shown that the aapT gene product exhibits both alpha-amylase and pullulanase activities with one active site. The optimum temperature and pH for enzyme activity were found to be 70 degrees C and pH 9, respectively. Furthermore, AapT was found to strongly adsorb to crystalline cellulose (Avicel) and raw corn starch. Final hydrolyzed products from soluble starch range from maltose (G2) to maltotetraose (G4). Only maltotriose (G3) was produced from pullulan. The enzyme also hydrolyzes raw starch under a broad range of conditions (60 to 70 degrees C and pH 8 to 9).
Full text
PDF
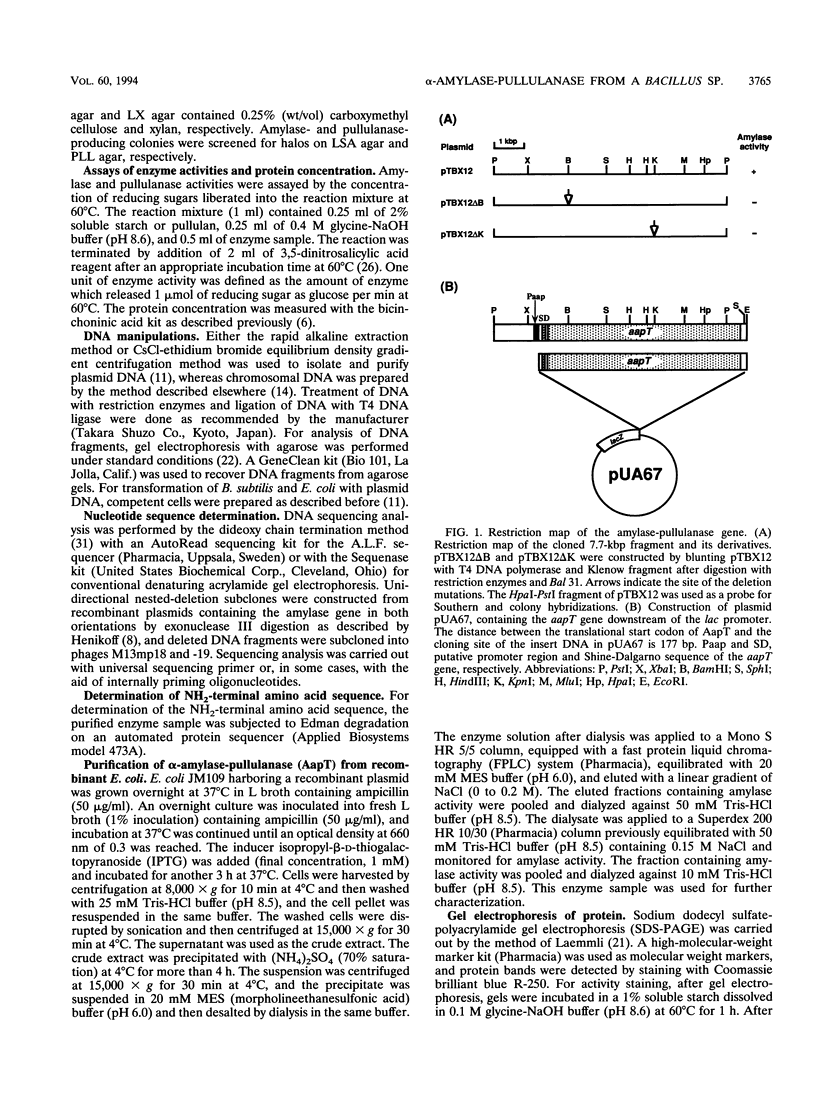



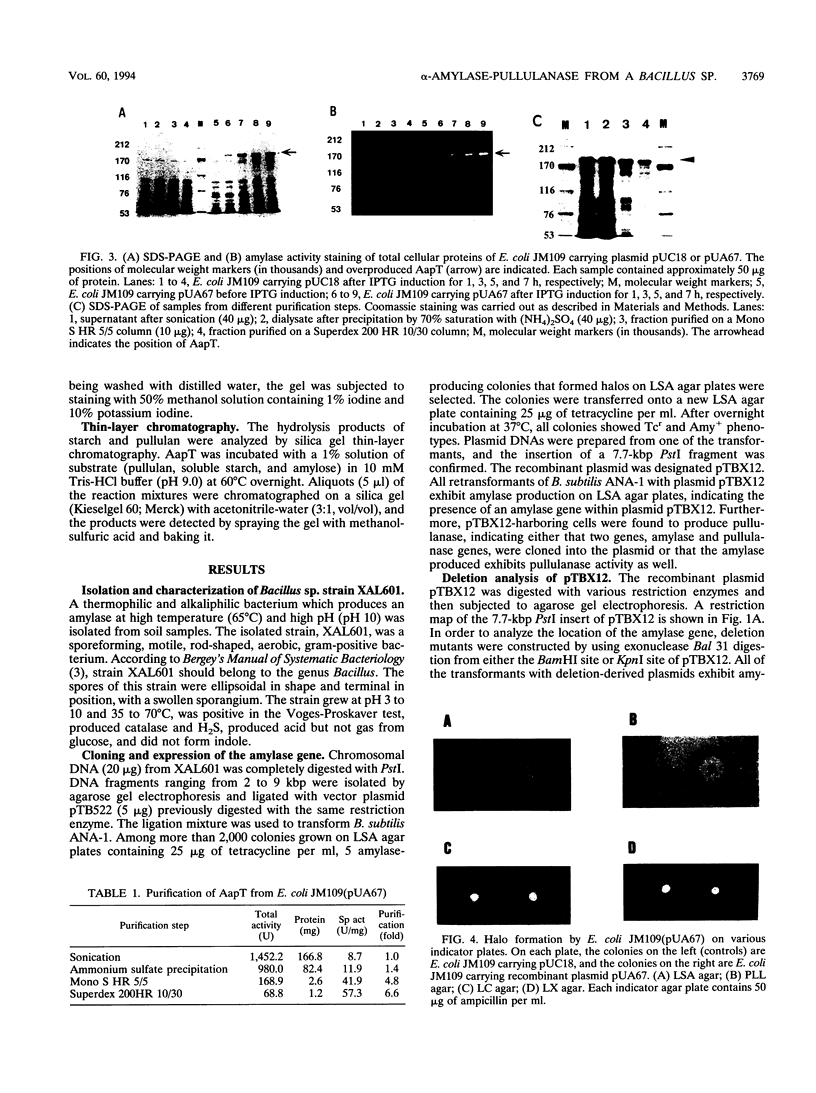


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boyer E. W., Ingle M. B. Extracellular alkaline amylase from a Bacillus species. J Bacteriol. 1972 Jun;110(3):992–1000. doi: 10.1128/jb.110.3.992-1000.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAMPBELL L. L., Jr Purification and properties of an alpha-amylase from facultative thermophilic bacteria. Arch Biochem Biophys. 1955 Jan;54(1):154–161. doi: 10.1016/0003-9861(55)90018-7. [DOI] [PubMed] [Google Scholar]
- Coleman R. D., Yang S. S., McAlister M. P. Cloning of the debranching-enzyme gene from Thermoanaerobium brockii into Escherichia coli and Bacillus subtilis. J Bacteriol. 1987 Sep;169(9):4302–4307. doi: 10.1128/jb.169.9.4302-4307.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S., Kakihara H., Woo K. B., Lejeune A., Kanemoto M., Sakaguchi K., Imanaka T. Cyclization characteristics of cyclodextrin glucanotransferase are conferred by the NH2-terminal region of the enzyme. Appl Environ Microbiol. 1992 Dec;58(12):4016–4025. doi: 10.1128/aem.58.12.4016-4025.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto T., Foong F., Shoseyov O., Doi R. H. Analysis of functional domains of endoglucanases from Clostridium cellulovorans by gene cloning, nucleotide sequencing and chimeric protein construction. Mol Gen Genet. 1992 Feb;231(3):472–479. doi: 10.1007/BF00292718. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Imanaka T., Fujii M., Aiba S. Isolation and characterization of antibiotic resistance plasmids from thermophilic bacilli and construction of deletion plasmids. J Bacteriol. 1981 Jun;146(3):1091–1097. doi: 10.1128/jb.146.3.1091-1097.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka T., Fujii M., Aramori I., Aiba S. Transformation of Bacillus stearothermophilus with plasmid DNA and characterization of shuttle vector plasmids between Bacillus stearothermophilus and Bacillus subtilis. J Bacteriol. 1982 Mar;149(3):824–830. doi: 10.1128/jb.149.3.824-830.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka T., Kuriki T. Pattern of action of Bacillus stearothermophilus neopullulanase on pullulan. J Bacteriol. 1989 Jan;171(1):369–374. doi: 10.1128/jb.171.1.369-374.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka T., Shibazaki M., Takagi M. A new way of enhancing the thermostability of proteases. Nature. 1986 Dec 18;324(6098):695–697. doi: 10.1038/324695a0. [DOI] [PubMed] [Google Scholar]
- Imanaka T., Tanaka T., Tsunekawa H., Aiba S. Cloning of the genes for penicillinase, penP and penI, of Bacillus licheniformis in some vector plasmids and their expression in Escherichia coli, Bacillus subtilis, and Bacillus licheniformis. J Bacteriol. 1981 Sep;147(3):776–786. doi: 10.1128/jb.147.3.776-786.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura F., Doi R. H. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J Bacteriol. 1984 Oct;160(1):442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriki T., Imanaka T. Nucleotide sequence of the neopullulanase gene from Bacillus stearothermophilus. J Gen Microbiol. 1989 Jun;135(6):1521–1528. doi: 10.1099/00221287-135-6-1521. [DOI] [PubMed] [Google Scholar]
- Kuriki T., Okada S., Imanaka T. New type of pullulanase from Bacillus stearothermophilus and molecular cloning and expression of the gene in Bacillus subtilis. J Bacteriol. 1988 Apr;170(4):1554–1559. doi: 10.1128/jb.170.4.1554-1559.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriki T., Park J. H., Okada S., Imanaka T. Purification and Characterization of Thermostable Pullulanase from Bacillus stearothermophilus and Molecular Cloning and Expression of the Gene in Bacillus subtilis. Appl Environ Microbiol. 1988 Nov;54(11):2881–2883. doi: 10.1128/aem.54.11.2881-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriki T., Takata H., Okada S., Imanaka T. Analysis of the active center of Bacillus stearothermophilus neopullulanase. J Bacteriol. 1991 Oct;173(19):6147–6152. doi: 10.1128/jb.173.19.6147-6152.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Matsuura Y., Kusunoki M., Harada W., Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem. 1984 Mar;95(3):697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- Melasniemi H., Paloheimo M., Hemiö L. Nucleotide sequence of the alpha-amylase-pullulanase gene from Clostridium thermohydrosulfuricum. J Gen Microbiol. 1990 Mar;136(3):447–454. doi: 10.1099/00221287-136-3-447. [DOI] [PubMed] [Google Scholar]
- Melasniemi H. Purification and some properties of the extracellular alpha-amylase-pullulanase produced by Clostridium thermohydrosulfuricum. Biochem J. 1988 Mar 15;250(3):813–818. doi: 10.1042/bj2500813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasahara K., Imanishi A., Isemura T. Studies on thermophilic alpha-amylase from Bacillus stearothermophilus. I. Some general and physico-chemical properties of thermophilic alpha-amylase. J Biochem. 1970 Jan;67(1):65–75. doi: 10.1093/oxfordjournals.jbchem.a129235. [DOI] [PubMed] [Google Scholar]
- Saha B. C., Lamed R., Lee C. Y., Mathupala S. P., Zeikus J. G. Characterization of an endo-Acting Amylopullulanase from Thermoanaerobacter Strain B6A. Appl Environ Microbiol. 1990 Apr;56(4):881–886. doi: 10.1128/aem.56.4.881-886.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata H., Kuriki T., Okada S., Takesada Y., Iizuka M., Minamiura N., Imanaka T. Action of neopullulanase. Neopullulanase catalyzes both hydrolysis and transglycosylation at alpha-(1----4)- and alpha-(1----6)-glucosidic linkages. J Biol Chem. 1992 Sep 15;267(26):18447–18452. [PubMed] [Google Scholar]
- Wong W. K., Gerhard B., Guo Z. M., Kilburn D. G., Warren A. J., Miller R. C., Jr Characterization and structure of an endoglucanase gene cenA of Cellulomonas fimi. Gene. 1986;44(2-3):315–324. doi: 10.1016/0378-1119(86)90196-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zhang M., Nakai H., Imanaka T. Useful Host-Vector Systems in Bacillus stearothermophilus. Appl Environ Microbiol. 1988 Dec;54(12):3162–3164. doi: 10.1128/aem.54.12.3162-3164.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]