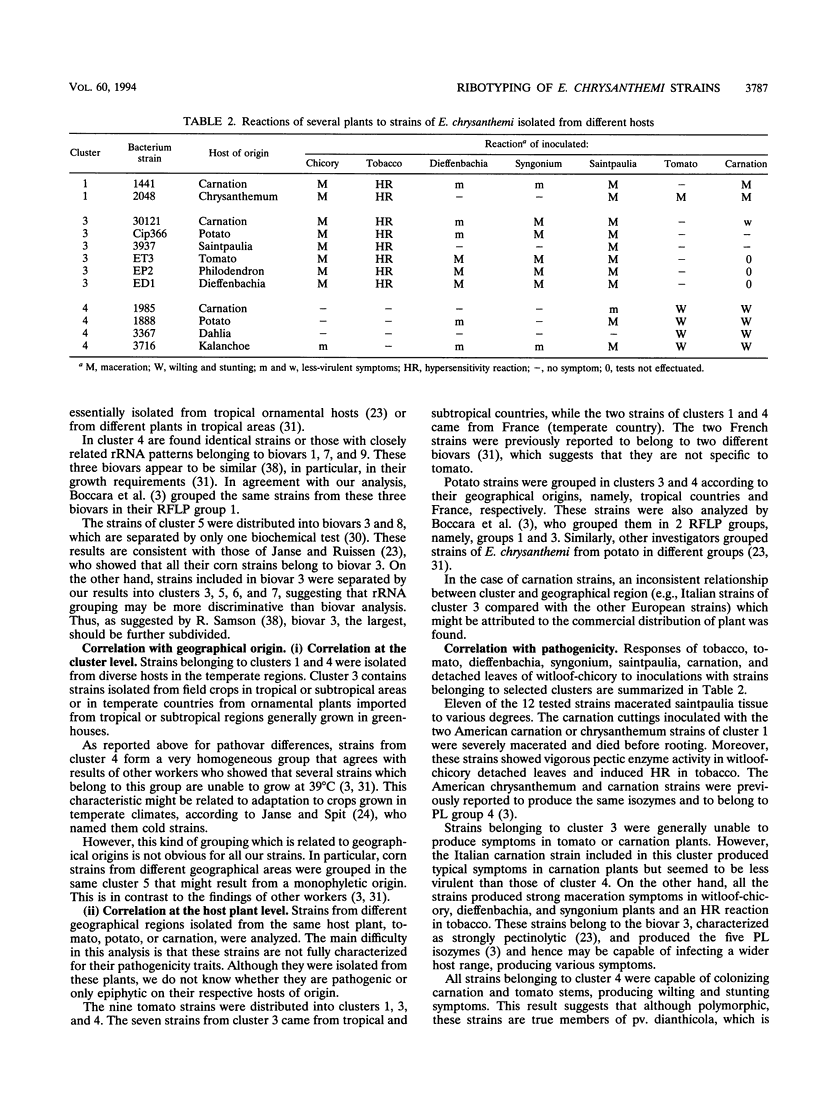
Abstract
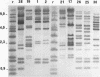
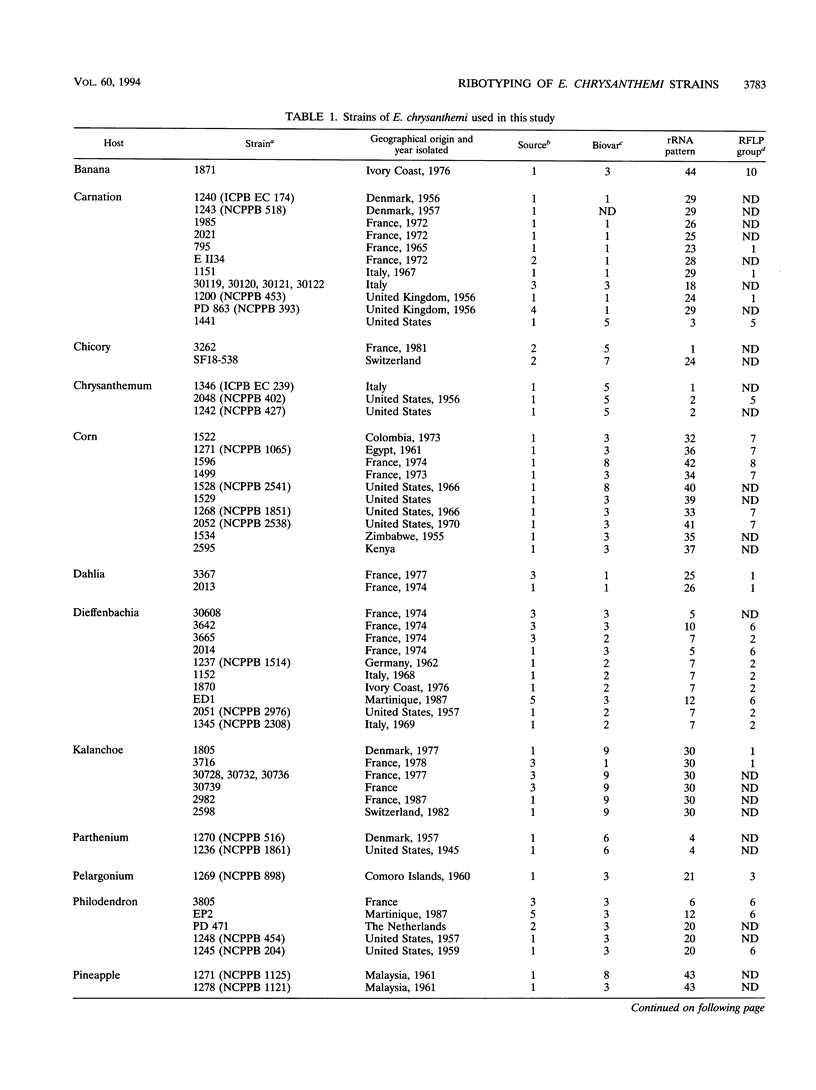
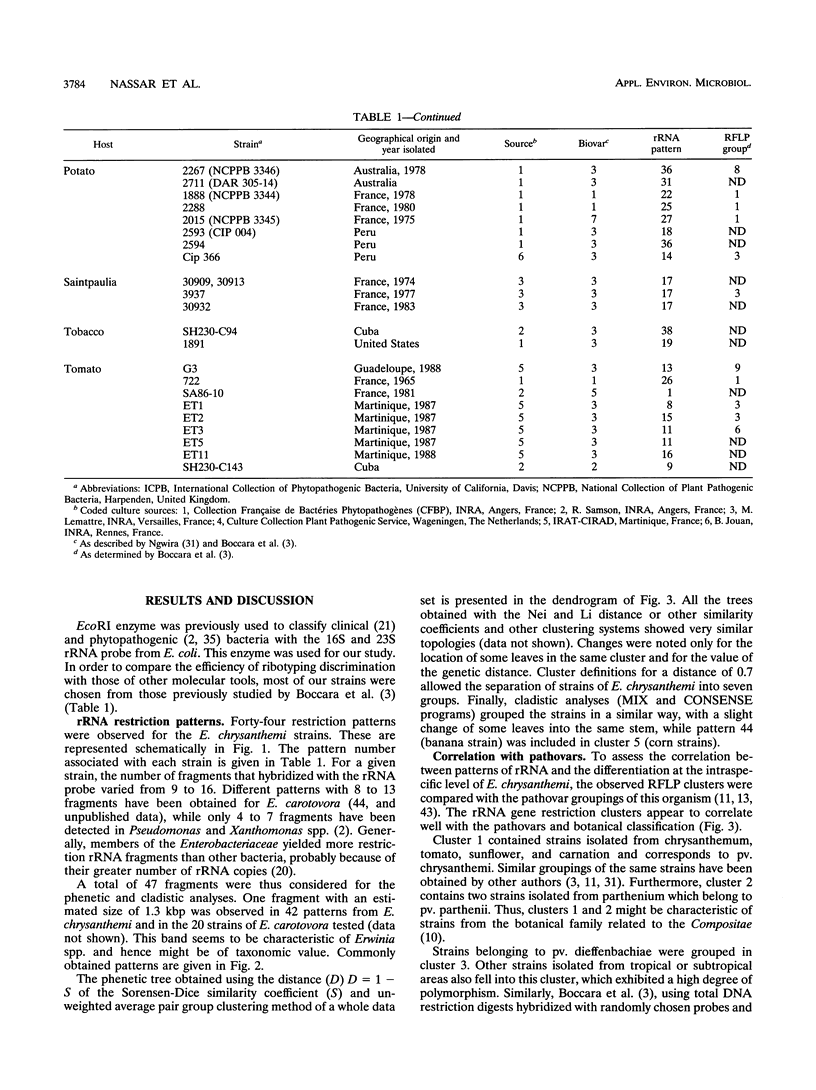
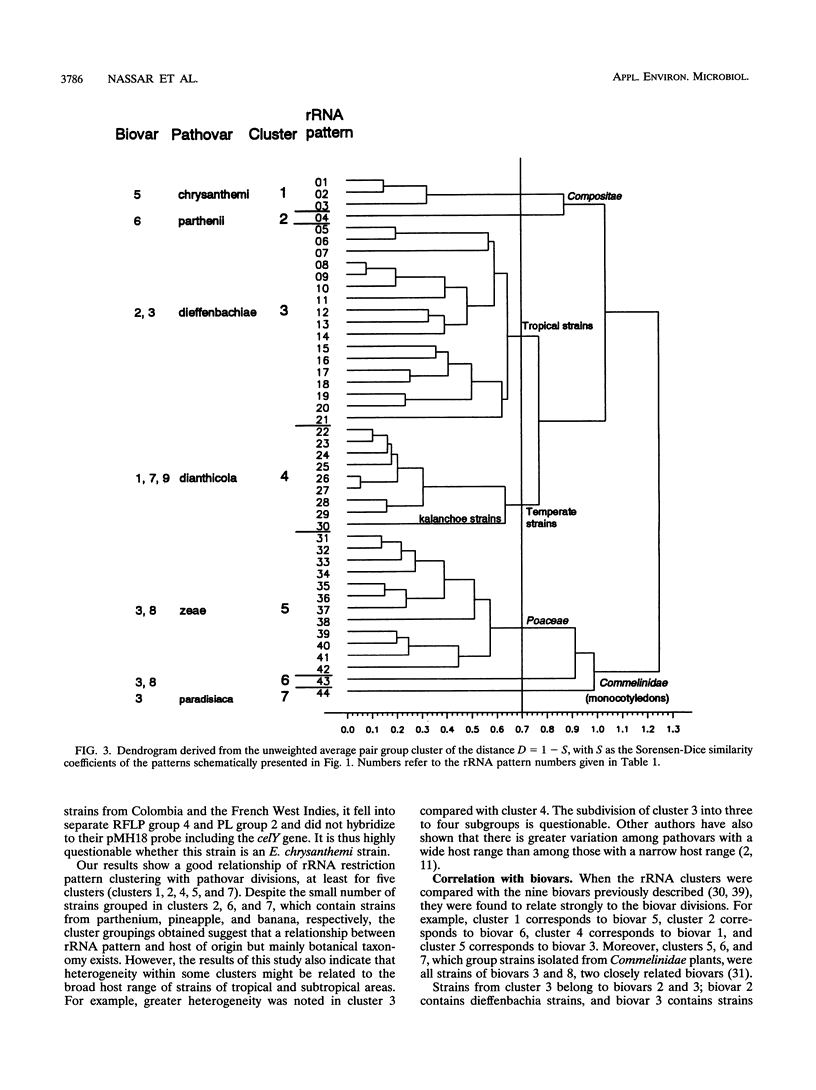
16S and 23S rRNAs from Escherichia coli were used to study the relationship among a representative collection of strains of Erwinia chrysanthemi differing in their original host and geographical origin. Phenetic analysis of restriction fragment length polymorphisms allowed the distribution of the studied strains into seven clusters. These clusters were similar to those obtained by cladistic methods and appeared to correlate well with the established pathovars and biovars but to a lesser extent with geographical distribution. Except for two groups of strains defined as tropical and temperate isolates (clusters 3 and 4, respectively), our clustering correlated well with botanical classifications of host plants. However, the rRNA groupings were shown to be more discriminative than biovar analysis. To assess the relationship between rRNA clusters and pathogenicity, 12 representative strains from different clusters were tested for pathogenicity on different plants. The two typical symptoms, maceration and wilting, were observed for these strains. The occurrence of the tobacco hypersensitivity reaction for a subset of these strains is discussed in light of recent results concerning the presence of an hrp gene. Considering symptom expression only, rather than the capacity for plant infection, strains from the same cluster were shown to induce similar symptoms in test plants. Thus, since host specificity is still quite controversial, rRNA patterns may constitute a useful tool in taxonomic and epidemiological studies of Erwinia chrysanthemi species.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berthier Y., Verdier V., Guesdon J. L., Chevrier D., Denis J. B., Decoux G., Lemattre M. Characterization of Xanthomonas campestris Pathovars by rRNA Gene Restriction Patterns. Appl Environ Microbiol. 1993 Mar;59(3):851–859. doi: 10.1128/aem.59.3.851-859.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick R. D., Majerczak D. R., Coplin D. L. Erwinia stewartii WtsA, a positive regulator of pathogenicity gene expression, is similar to Pseudomonas syringae pv. phaseolicola HrpS. Mol Microbiol. 1993 Aug;9(3):477–485. doi: 10.1111/j.1365-2958.1993.tb01709.x. [DOI] [PubMed] [Google Scholar]
- Grimm C., Panopoulos N. J. The predicted protein product of a pathogenicity locus from Pseudomonas syringae pv. phaseolicola is homologous to a highly conserved domain of several procaryotic regulatory proteins. J Bacteriol. 1989 Sep;171(9):5031–5038. doi: 10.1128/jb.171.9.5031-5038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimont F., Chevrier D., Grimont P. A., Lefevre M., Guesdon J. L. Acetylaminofluorene-labelled ribosomal RNA for use in molecular epidemiology and taxonomy. Res Microbiol. 1989 Sep;140(7):447–454. doi: 10.1016/0923-2508(89)90065-x. [DOI] [PubMed] [Google Scholar]
- Grimont F., Grimont P. A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- Klotz L. C., Zimm B. H. Size of DNA determined by viscoelastic measurements: results on bacteriophages, Bacillus subtilis and Escherichia coli. J Mol Biol. 1972 Dec 30;72(3):779–800. doi: 10.1016/0022-2836(72)90191-x. [DOI] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried J. L., Collmer A. Comparison of pectic enzymes produced by Erwinia chrysanthemi, Erwinia carotovora subsp. carotovora, and Erwinia carotovora subsp. atroseptica. Appl Environ Microbiol. 1986 Aug;52(2):305–310. doi: 10.1128/aem.52.2.305-310.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Starr M. P., Mandel M. DNA base composition and taxonomy of phyopathogenic and other enterobacteria. J Gen Microbiol. 1969 Apr;56(1):113–123. doi: 10.1099/00221287-56-1-113. [DOI] [PubMed] [Google Scholar]