Abstract
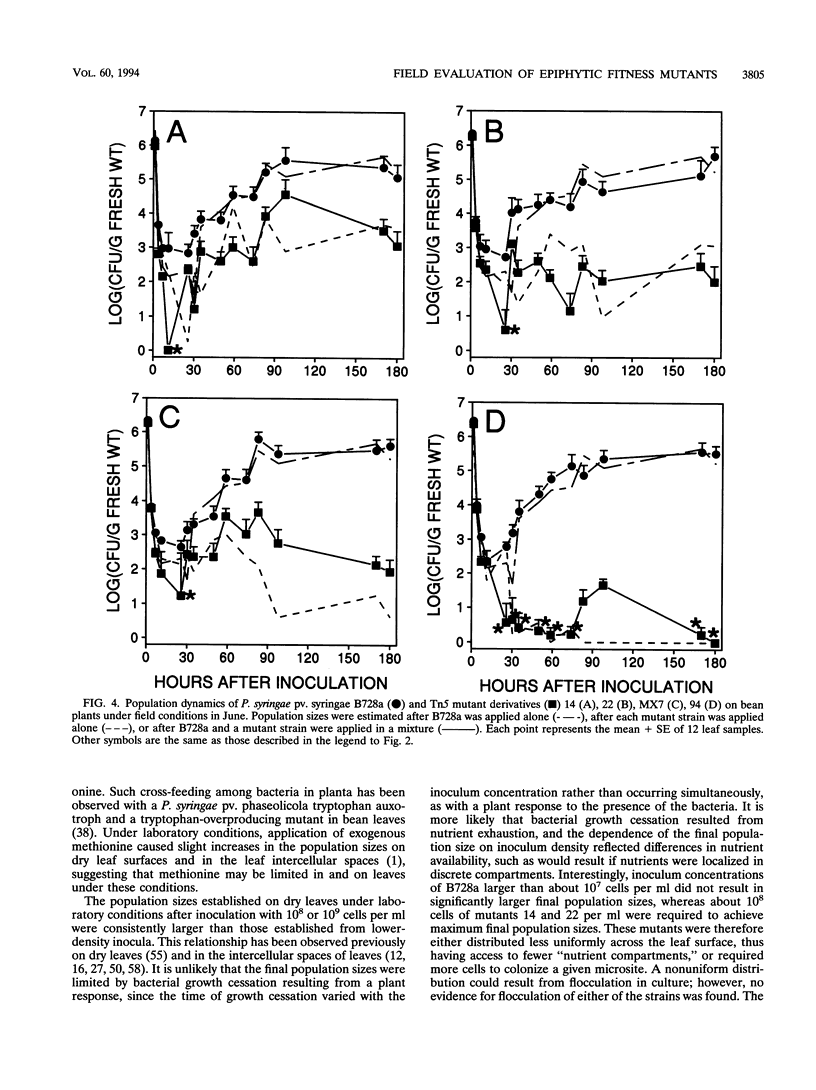
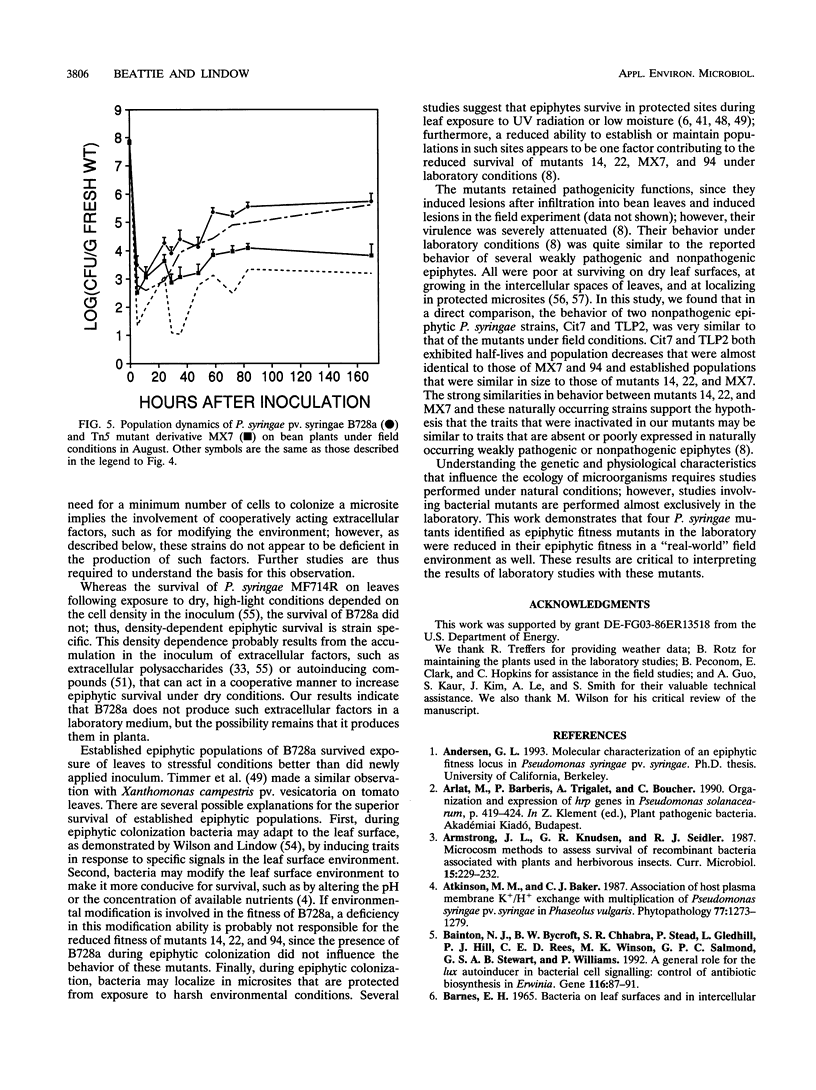
The epiphytic fitness of four Tn5 mutants of Pseudomonas syringae that exhibited reduced epiphytic fitness in the laboratory was evaluated under field conditions. The mutants differed more from the parental strain under field conditions than under laboratory conditions in their survival immediately following inoculation onto bean leaves and in the size of the epiphytic populations that they established, demonstrating that their fitness was reduced more under field conditions than in the laboratory. Under both conditions, the four mutants exhibited distinctive behaviors. One mutant exhibited particularly large population decreases and short half-lives following inoculation but grew epiphytically at near-wild-type rates, while the others exhibited reduced survival only in the warmest, driest conditions tested and grew epiphytically at reduced rates or, in the case of one mutant, not at all. The presence of the parental strain, B728a, did not influence the survival or growth of three of the mutants under field conditions; however, one mutant, an auxotroph, established larger populations in the presence of B728a than in its absence, possibly because of cross-feeding by B728a in planta. Experiments with B728a demonstrated that established epiphytic populations survived exposure of leaves to dry conditions better than newly inoculated cells did and that epiphytic survival was not dependent on the cell density in the inoculum. Three of the mutants behaved similarly to two nonpathogenic strains of P. syringae, suggesting that the mutants may be altered in traits that are missing or poorly expressed in naturally occurring nonpathogenic epiphytes.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bainton N. J., Bycroft B. W., Chhabra S. R., Stead P., Gledhill L., Hill P. J., Rees C. E., Winson M. K., Salmond G. P., Stewart G. S. A general role for the lux autoinducer in bacterial cell signalling: control of antibiotic biosynthesis in Erwinia. Gene. 1992 Jul 1;116(1):87–91. doi: 10.1016/0378-1119(92)90633-z. [DOI] [PubMed] [Google Scholar]
- Barnes E. H. Bacteria on Leaf Surfaces and in Intercellular Leaf Spaces. Science. 1965 Mar 5;147(3662):1151–1152. doi: 10.1126/science.147.3662.1151. [DOI] [PubMed] [Google Scholar]
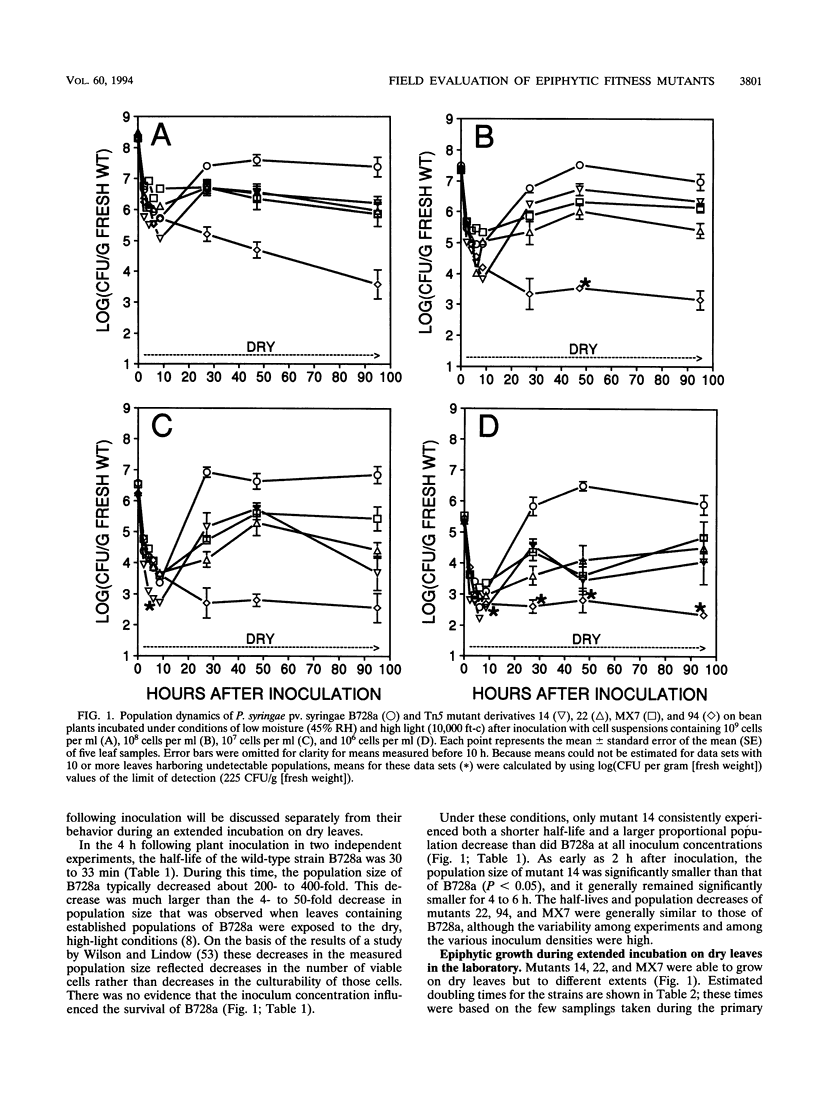
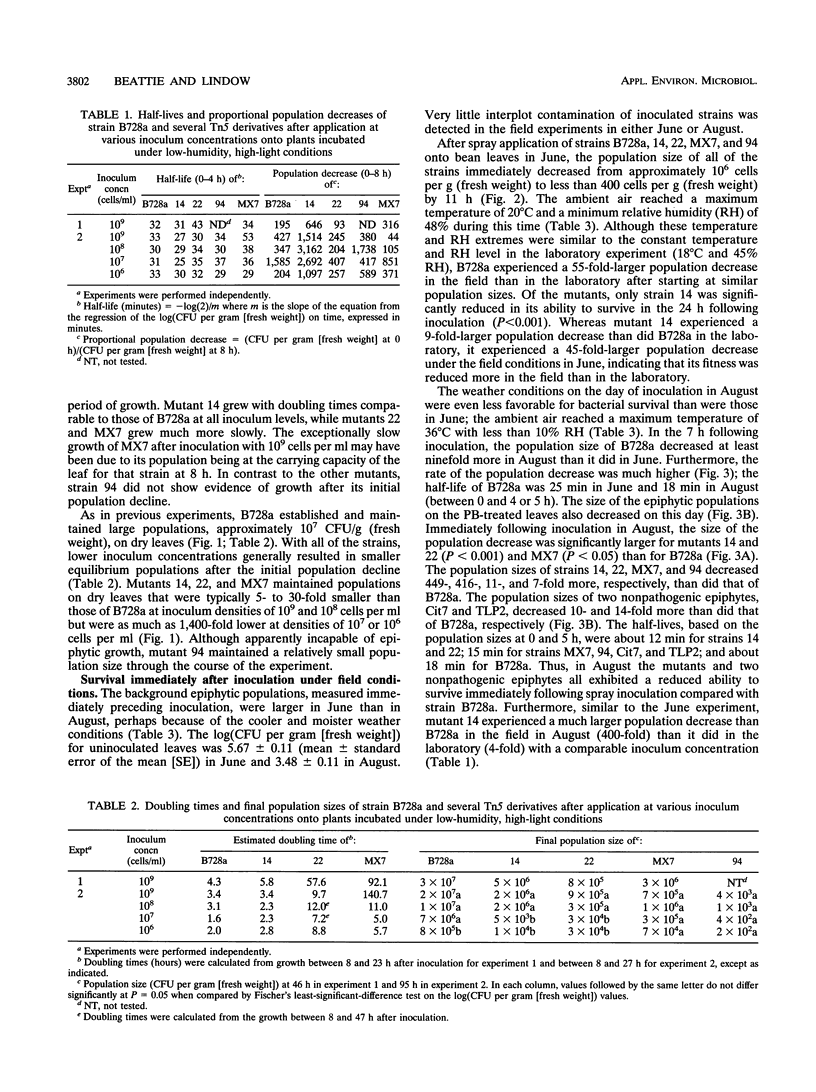
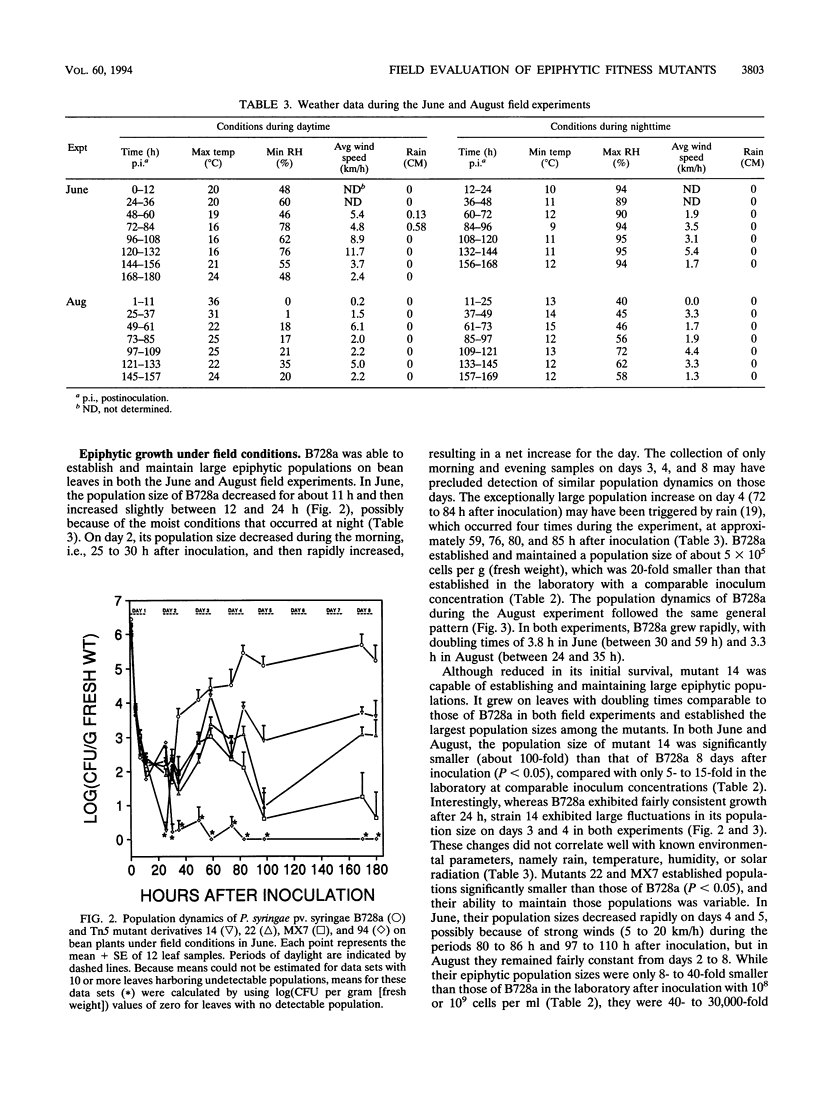
- Beattie G. A., Lindow S. E. Survival, Growth, and Localization of Epiphytic Fitness Mutants of Pseudomonas syringae on Leaves. Appl Environ Microbiol. 1994 Oct;60(10):3790–3798. doi: 10.1128/aem.60.10.3790-3798.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu C., Van Gijsegem F. Identification of plant-inducible genes in Erwinia chrysanthemi 3937. J Bacteriol. 1990 Mar;172(3):1569–1575. doi: 10.1128/jb.172.3.1569-1575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke H. R., Leigh J. A., Douglas C. J. Molecular signals in the interactions between plants and microbes. Cell. 1992 Oct 16;71(2):191–199. doi: 10.1016/0092-8674(92)90348-g. [DOI] [PubMed] [Google Scholar]
- Dénarié J., Cullimore J. Lipo-oligosaccharide nodulation factors: a minireview new class of signaling molecules mediating recognition and morphogenesis. Cell. 1993 Sep 24;74(6):951–954. doi: 10.1016/0092-8674(93)90717-5. [DOI] [PubMed] [Google Scholar]
- Eberhard A., Burlingame A. L., Eberhard C., Kenyon G. L., Nealson K. H., Oppenheimer N. J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981 Apr 28;20(9):2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- Harter H. L., Moore A. H. Iterative maximum-likelihood estimation of the parameters of normal populations from singly and doubly censored samples. Biometrika. 1966 Jun;53(1):205–213. [PubMed] [Google Scholar]
- Hirano S. S., Nordheim E. V., Arny D. C., Upper C. D. Lognormal distribution of epiphytic bacterial populations on leaf surfaces. Appl Environ Microbiol. 1982 Sep;44(3):695–700. doi: 10.1128/aem.44.3.695-700.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Beppu T. Autoregulatory factors of secondary metabolism and morphogenesis in actinomycetes. Crit Rev Biotechnol. 1990;10(3):191–204. doi: 10.3109/07388559009038207. [DOI] [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- Kamoun S., Kado C. I. A plant-inducible gene of Xanthomonas campestris pv. campestris encodes an exocellular component required for growth in the host and hypersensitivity on nonhosts. J Bacteriol. 1990 Sep;172(9):5165–5172. doi: 10.1128/jb.172.9.5165-5172.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemoto R. H., Powell A. T., Akiyoshi D. E., Regier D. A., Kerstetter R. A., Nester E. W., Hawes M. C., Gordon M. P. Nucleotide sequence and analysis of the plant-inducible locus pinF from Agrobacterium tumefaciens. J Bacteriol. 1989 May;171(5):2506–2512. doi: 10.1128/jb.171.5.2506-2512.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren P. B., Frederick R., Govindarajan A. G., Panopoulos N. J., Staskawicz B. J., Lindow S. E. An ice nucleation reporter gene system: identification of inducible pathogenicity genes in Pseudomonas syringae pv. phaseolicola. EMBO J. 1989 May;8(5):1291–1301. doi: 10.1002/j.1460-2075.1989.tb03508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow S. E., Andersen G., Beattie G. A. Characteristics of Insertional Mutants of Pseudomonas syringae with Reduced Epiphytic Fitness. Appl Environ Microbiol. 1993 May;59(5):1593–1601. doi: 10.1128/aem.59.5.1593-1601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow S. E. Novel method for identifying bacterial mutants with reduced epiphytic fitness. Appl Environ Microbiol. 1993 May;59(5):1586–1592. doi: 10.1128/aem.59.5.1586-1592.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson K. H., Platt T., Hastings J. W. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol. 1970 Oct;104(1):313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn A. E., Barber C. E., Daniels M. J. Identification of plant-induced genes of the bacterial pathogen Xanthomonas campestris pathovar campestris using a promoter-probe plasmid. EMBO J. 1987 Jan;6(1):23–28. doi: 10.1002/j.1460-2075.1987.tb04713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper K. R., Beck von Bodman S., Farrand S. K. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature. 1993 Apr 1;362(6419):448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- Rahme L. G., Mindrinos M. N., Panopoulos N. J. Genetic and transcriptional organization of the hrp cluster of Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1991 Jan;173(2):575–586. doi: 10.1128/jb.173.2.575-586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P. J., Whitcombe D., Wharam S., Gibson M., Allison G., Bunce N., Barallon R., Douglas P., Mulholland V., Stevens S. Molecular cloning and characterization of 13 out genes from Erwinia carotovora subspecies carotovora: genes encoding members of a general secretion pathway (GSP) widespread in gram-negative bacteria. Mol Microbiol. 1993 May;8(3):443–456. doi: 10.1111/j.1365-2958.1993.tb01589.x. [DOI] [PubMed] [Google Scholar]
- Williams P., Bainton N. J., Swift S., Chhabra S. R., Winson M. K., Stewart G. S., Salmond G. P., Bycroft B. W. Small molecule-mediated density-dependent control of gene expression in prokaryotes: bioluminescence and the biosynthesis of carbapenem antibiotics. FEMS Microbiol Lett. 1992 Dec 15;100(1-3):161–167. doi: 10.1111/j.1574-6968.1992.tb14035.x. [DOI] [PubMed] [Google Scholar]
- Wilson M., Lindow S. E. Effect of phenotypic plasticity on epiphytic survival and colonization by Pseudomonas syringae. Appl Environ Microbiol. 1993 Feb;59(2):410–416. doi: 10.1128/aem.59.2.410-416.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M., Lindow S. E. Relationship of total viable and culturable cells in epiphytic populations of Pseudomonas syringae. Appl Environ Microbiol. 1992 Dec;58(12):3908–3913. doi: 10.1128/aem.58.12.3908-3913.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]



