Abstract
When demand for cholesterol rises in mammalian cells, the sterol regulatory element (SRE) binding proteins (SREBPs) are released from their membrane anchor through proteolysis. Then, the N-terminal region enters the nucleus and activates genes of cholesterol uptake and biosynthesis. Basic helix–loop–helix (bHLH) proteins such as SREBPs bind to a palindromic DNA sequence called the E-box (5′-CANNTG-3′). However, SREBPs are special because they also bind direct repeat elements called SREs. Importantly, sterol regulation of all promoters studied thus far is mediated by SREBP binding only to SREs. To study the reason for this we converted the direct repeat SRE from the sterol-regulated low-density lipoprotein receptor promoter into an E-box. In this report we show that SREBPs are still able to bind and activate this promoter however, sterol regulation is lost. The results are consistent with the mutant promoter being a target for promiscuous activation by constitutively expressed E-box binding bHLH proteins that are not regulated by cholesterol. Kim and coworkers [Kim, J. B., Spotts, G. D., Halvorsen, Y.-D., Shih, H.-M., Ellenberger, T., Towle, H. C. & Spiegelman, B. M. (1995) Mol. Cell. Biol. 15, 2582–2588] demonstrated that the dual DNA binding specificity of SREBPs is caused by a specific tyrosine in the conserved basic region of the DNA binding domain that corresponds to an arginine in all other bHLH proteins that recognize only E-boxes. Taken together the data suggest an evolutionary mechanism where a DNA binding protein along with its recognition site have coevolved to ensure maximal specificity and sensitivity in a crucial nutritional regulatory response.
When the sterol level in mammalian cells declines the genes for proteins of cholesterol uptake and biosynthesis are up-regulated (1, 2). The central proteins responsible for maintaining cholesterol homeostasis are the sterol regulatory element binding proteins (SREBPs) (3). They are a family of transcription factors that bind to the sterol regulatory element (SRE) in the promoters of target genes and activate their transcription in a cholesterol dependent fashion. The first SRE element that was identified with single nucleotide accuracy was the sequence 5′-ATCACCCCAC-3′ in repeat 2 of the human low density lipoprotein (LDL) receptor promoter, which contains a direct repeat (5′-PyCCAC-3′) separated by a nonessential base. The functionally identified SRE elements in other sterol-regulated promoters are all similar to this element but there can be individual base differences with the LDL receptor site (4–10). Thus, the full range of potential SRE sites that can direct sterol regulation in the context of specific promoters has not been firmly established.
The SREBP family is comprised of several related proteins, at least two isoforms of SREBP-1 are expressed from a single gene and a separate gene encodes the singular SREBP-2 protein (3, 11, 12). In cultured cells the SREBPs are coordinately regulated by cellular sterol levels by a two-step proteolytic mechanism. When sterol levels are high the primary translation product is functionally a precursor that is anchored to the membrane of the endoplasmic reticulum in a hairpin fashion via two transmembrane spans (13, 14). When intracellular sterol levels decrease the full-length SREBPs are proteolytically processed in two steps (15) ultimately releasing the transcriptionally active N-terminal half of the proteins that translocate to the nucleus and activate transcription via binding to SREs contained within promoters of specific target genes (3).
Although SREBPs are the key transcriptional regulatory proteins that maintain sterol balance, in all promoters that have been carefully studied a generic coactivating transcription factor and its associated binding site are also required for regulated expression (16–18). For example, in the LDL receptor promoter this coactivator is the ubiquitous transcription factor Sp1 (18); for hydroxymethylglutaryl-CoA synthase and farnesyl diphosphate synthase, the generic factor is nuclear factor-Y (19, 20).
Although SREBPs were first identified as proteins that regulate intracellular cholesterol balance, subsequent studies have demonstrated their involvement in the regulation of fatty acid metabolism as well (7, 21–23). Variant SRE sites are present in the promoter regions for acetyl-CoA carboxylase, fatty acid synthase (FAS), and glycerol-3-phosphate acyltransferase, which are genes of fatty acid metabolism (7, 21, 22).
The SREBPs belong to the basic helix–loop–helix (bHLH) leucine zipper class of transcription factors that includes Myc, Max, and upstream stimulatory factor (USF) (24). bHLH proteins bind to a palindromic consensus DNA sequence known as the E-box (5′-CANNTG-3′) with high affinity and mediate transcriptional activation through the target E-box(es) in the promoters of selective genes. A more detailed investigation of their DNA binding specificity has shown that bHLH leucine zipper-containing proteins can bind and activate transcription through nonconsensus sites as well (11, 25).
A classic example of non-E-box DNA recognition for bHLH leucine zipper proteins is provided by the SREBPs because they bind to a palindromic E-box as well as to the direct repeat SRE elements such as that found in the promoter for the LDL receptor (11, 26). Spiegelman and colleagues showed that the ability of SREBPs to bind both SREs and E-boxes is caused largely by a critical tyrosine residue located at a position in the basic DNA binding domain that is occupied by an arginine residue in other bHLH proteins that typically recognize E-box sites (26).
Interestingly, although SREBPs bind to the palindromic E-box with high affinity in vitro, sterol regulation of all promoters through SREBP action described thus far is mediated through SREs and not E-boxes (4, 6–10, 27–30). Thus, even though an E-box is contained within the region required for sterol regulation of the FAS promoter the E-box element itself was not responsible. Rather, two SREBP sites each containing half of the E-box were shown to be the targets for SREBP binding and sterol regulation (9). There are two simple reasons why sterol regulation by SREBP may not occur through its recognition of E-box target sites. (i) It was formally possible that although SREBPs bind E-boxes, the resulting protein-DNA conformation is incompatible with transcriptional activation. (ii) Alternatively, and a more likely explanation was that SREBPs do not utilize E-box targets for sterol regulation because it would result in a loss in specificity. E-box sites would be recognized by other bHLH proteins that are constitutively expressed and not dependent on sterol-regulated processing to accumulate within the nucleus. Thus, E-boxes within promoters for potentially sterol-regulated genes would be activated indiscriminately regardless of the sterol level of the cell. When new cholesterol is required and SREBP translocates to the nucleus the added activation mediated by SREBP would be of minimal significance and the net difference in gene expression under low and high sterol levels would be negligible.
We wanted to test these hypotheses directly so we converted the SRE of a natural sterol-regulated promoter into a palindromic E-box by introducing a minimal number of base pair changes into a simple well-defined SRE. Thus, three base pair changes were introduced into the SRE element of the LDL receptor promoter converting it from a direct repeat SRE into a palindromic E-box. Then we compared the mutant relative to the wild-type promoter for SREBP binding, sterol regulation, and activation by both ectopically expressed SREBP-1a and a hybrid USF2-VP16 activator that only binds and activates transcription through palindromic E-box sites.
The data demonstrated that SREBP can bind and activate transcription through the E-box, however, sterol regulation was lost because the E-box containing mutant promoter was significantly activated under conditions where the concentration of cellular sterols is high and thus the nuclear content of SREBPs is minimal. As expected the E-box containing mutant, but not the wild type, was efficiently activated by the USF2-VP16 activator (31). These studies suggest that sterol-regulated promoters and their corresponding specific transcription factors, the SREBPs, have coevolved to achieve optimal specificity for precise metabolic regulation.
MATERIALS AND METHODS
Plasmids.
The construction of the reporter plasmid for the wild-type LDL receptor gene has been described (18). The mutant low density lipoprotein E-box (LDLE)-containing promoter that has an E-box motif in place of the direct repeat SRE was constructed by PCR amplification using the wild-type LDL receptor reporter plasmid as the template. A specific primer was designed containing a NheI site at its terminus that hybridized to the coding strand of the human LDL receptor promoter encompassing repeats 2 and 3 with a 3 base pair change from the wild-type sequence in repeat 2 to introduce the mutation shown in Fig. 1A. This was used together with a primer containing a SacI site engineered at its end that hybridized specifically to the noncoding strand corresponding to the pGL2 sequence (Promega) upstream of the LDL receptor promoter insert. The PCR product was then digested with NheI and SacI that flank the LDL receptor promoter insert, the DNA was gel purified and ligated to the large fragment of NheI and SacI digested LDL receptor wild-type promoter plasmid to yield LDLE-Luc. The integrity of the insert and the correct insertion of mutant bases was confirmed by DNA sequence analysis. The plasmid cytomegalovirus-USF2-VP16 plasmid (31) was a gift from H. Towle (University of Minnesota, Minneapolis) and all other plasmids have been described (32).
Figure 1.
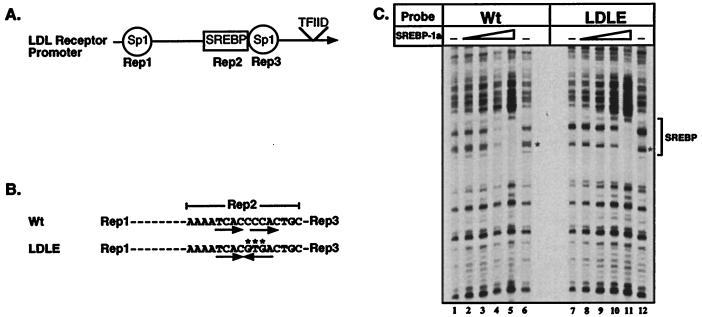
Wild-type and mutant LDL receptor promoters: Structure and DNase I footprint analysis by SREBP-1a. A schematic view of the region of the LDL receptor promoter required for sterol regulation containing repeats 1–3 (A) and a more detailed view of the sequence of the wild-type (LDL) and mutant repeat 2 (LDLE) are shown (B). The mutated bases are indicated by ∗ and the direct repeat and palindromic motifs are indicated by the arrows. (C). DNA probes end-labeled with 32P on the top strand were prepared from plasmids containing the wild-type (Wt, lanes 1–6) or mutant (LDLE, lanes 7–12) promoters. DNA probe (0.01 pmol) were used in each binding reaction in the absence or presence of increasing amounts (0.02, 0.06, 0.2, and 0.6 pmol) of recombinant SREBP-1a (amino acids 1–490). DNase I footprinting was performed as described in Materials and Methods. The asterisk on the autoradiogram (C) indicates the position of differences in the observed cleavage pattern of free DNA due to the 3-bp difference between the two probes. The minimal size of the DNase I footprint is indicated by the bracket at the right of the figure.
Cells and Media: Drosophila.
SL2 cells were cultured at 25°C in Shields and Sang Drosophila medium (Sigma) containing 10% (vol/vol) heat-inactivated fetal bovine serum. CV-1 cells were cultured in DMEM (Life Technologies) containing 10% (vol/vol) fetal bovine serum at 37°C and 8% CO2. HepG2 cells (American Type Culture Collection), were cultured in MEM (Life Technologies, Grand Island, NY) containing 10% fetal bovine serum (vol/vol) at 37°C and 5% CO2. Lipoprotein-deficient serum was prepared by ultra centrifugation as described (33). Stock solutions of cholesterol, and 25-hydroxycholesterol (Steraloids, Wilton, NH) were prepared in ethanol.
Transient DNA Transfections.
All transfections were performed by a standard calcium phosphate coprecipitation method as described before (18). Drosophila SL2 cells were plated at 1.2 × 106 cells per 60-mm dish and were transfected the following day and harvested 48 hr after transfection. HepG2 cells were plated at 1.75 × 105 cells per 60-mm dish and transfected the following day. Dishes were subjected to a glycerol shock 4–6 hr after transfection, rinsed three times with PBS, refed with normal media and incubated an additional 40–48 hr before harvest. CV1 cells were plated at 1.25 × 105 cells per 60-mm dish and transfected the following day. Eighteen hours after transfection the dishes were rinsed three times with PBS and duplicate dishes were fed with either induced media (DMEM containing 10% lipoprotein-depleted serum) or suppressed media (DMEM containing 10% lipoprotein-depleted serum supplemented with 10 μg/ml cholesterol and 1 μg/ml 25-hydroxycholesterol) and incubated an additional 24 hr and harvested as described above. Two dishes were used for each data point and cells from both dishes were combined during harvest and cell extracts were prepared by a standard freeze-thaw method in a 100-μl volume.
Enzyme Assays.
Luciferase activities were measured in a luminometer [Analytical Luminescence Laboratory (San Diego) Monolight 2010] using 20 μl of cell extract and 100 μl of luciferin reagent (Promega). The β-galactosidase assays were performed by using 50 μl of extract with a standard colorimetric assay containing 2-nitrophenyl β-d-galactopyranoside as the substrate (34). The luciferase activities in relative light units were divided by the β-galactosidase activities (A420/hr) for each sample.
Protein Purification.
The recombinant SREBP-1a protein (amino acids 1–490) was expressed in Escherichia coli as a fusion protein containing a 6× His-tag at the N terminus and purified by metal affinity chromatography as described (18). The concentration of the purified protein was assessed by SDS/PAGE followed by Coomassie blue staining.
DNase I Footprinting.
32P-end-labeled DNA probes were prepared from the indicated plasmids and incubated on ice in the absence or presence of purified recombinant SREBP-1a protein (amino acids 1–490), digested with DNase I, and processed further as described (18).
RESULTS
Although SREBPs are capable of binding E-boxes with high affinity in vitro (26, 30), sterol regulation of all carefully examined promoters so far is mediated through SREBP binding to SRE elements and not E-boxes (4, 6–10, 27–30). Therefore, it was possible that although SREBPs can recognize E-boxes in vitro, the resulting protein-DNA complex may not be properly oriented to facilitate interactions with required cofactors and/or components of the basal transcription machinery that are critical for transcriptional activation. To determine whether SREBPs were capable of activating an E-box-containing promoter in cells and whether it required the interaction of required coregulatory DNA binding proteins as is observed for SRE containing promoters (18), we designed a reporter construct containing three high-affinity E-boxes adjacent to a single Sp1 site and analyzed its expression in a cotransfection experiment in SL2 cells along with an added vector expressing SREBP-1a. An analogous reporter construct containing three SREs and a single Sp1 site described previously (18) was also included alongside for comparison. We found that both reporter constructs are efficiently activated by SREBP-1a thus SREBP-1a is indeed capable of mediating transcription through the E-box as well as through SRE-like sites (data not shown). This is consistent with supportive data from others that suggests SREBPs can activate transcription through E-box recognition (26, 35).
Although this experiment demonstrated that SREBPs can bind and activate transcription through the E-box the question of why E-box targets are not present in sterol regulatory DNA elements remained unresolved. We hypothesized that perhaps SRE targets were important not necessarily for activation per se but for specificity in sterol regulation. If sterol-regulated genes such as the LDL receptor were activated through E-box targets instead of SREs then perhaps sterol regulation would be minimal or nondetectable because other bHLH proteins (such as USF) that are expressed and targeted to the nucleus constitutively would bind to the E-boxes and activate gene expression even when sterol levels are relatively high.
To test this hypothesis in the context of a natural promoter, we utilized the native human LDL receptor promoter that appears to have a simple ordered structure of two Sp1 sites flanking a single direct repeat SRE element (Fig. 1A). We noted that by altering only three bases the direct repeat would be converted into an E-box that would be predicted to still bind SREBPs with high affinity (this mutant promoter is referred to as LDLE in Fig. 1B and throughout the text).
Before evaluating the mutant LDL receptor promoter for sterol regulation, we first tested its ability to bind SREBPs in vitro and to support activation by ectopically expressed SREBPs in transfected cells. The results of a DNase I footprint analysis for the binding of recombinant SREBP-1a on the wild-type (Wt) and mutant (LDLE) LDL receptor promoters is displayed in Fig. 1C. The results demonstrate that SREBP-1a bound similarly to the wild-type and E-box mutant (Fig. 1C). The data suggest that binding affinity of SREBP to the direct repeat SRE may be slightly higher than to the E-box (compare lanes 4 and 5 with 10 and 11). Although quantitative measurements of SREBP binding by the electorophoretic mobility shift assay suggests that both sites bind SREBP-1a with similar affinity (30).
Because SREBP-1a bound to the E-box mutant promoter similarly to the wild-type LDL receptor promoter, we compared the ability of SREBP-1a to activate the mutant and wild-type promoters in a cotransfection assay in Drosophila SL2 cells. We have exploited the SL2 system to evaluate the synergistic transcriptional activation by members of the SREBP and Sp1 families of transcriptional activators. These cells are devoid of endogenous active SREBP or Sp1 like activities but they support a robust synergistic activation when promoters that require both SREBP and Sp1 are transfected along with constructs designed to express both proteins.
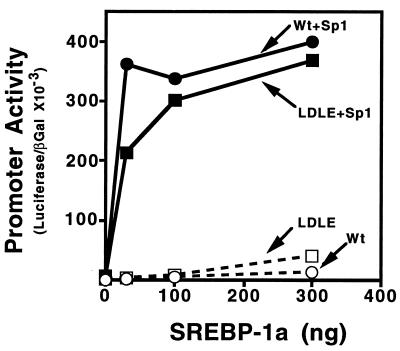
When increasing amounts of an SREBP-1a expression vector was transfected along with a constant amount of plasmid expressing Sp1 into SL2 cells both the wild-type and mutant LDL receptor promoters were activated in a dose-dependent fashion (Fig. 2). However, consistent with our earlier studies (18, 32, 36), neither promoter was significantly activated by SREBP-1a in the absence of Sp1. These data indicate that not only is the mutant promoter capable of responding to SREBPs but that activation through an E-box is also dependent on synergistic interactions with the Sp1 transcription factor that binds to a neighboring site. Thus, SREBP activation through an E-box requires a coregulatory DNA binding factor just as it does when it activates through an SRE element (18).
Figure 2.
Activation of the wild-type and mutant LDL receptor promoter by SREBP-1a and Sp1. Drosophila SL2 cells were transfected with increasing amounts of pPACSREBP-1a DNA alone (□, ○) or in the presence (▪, •) of a constant amount (25 ng/60-mm dish) of pPACSp1 expression vectors as indicated. The wild-type (Wt; •, ○) or mutant (LDLE; ▪, □) LDL receptor promoter fused to the luciferase gene (2 μg per 60-mm dish) were used as the reporters. Transfection, harvesting, preparation of cell extracts, and enzyme assays were performed as described in Materials and Methods. A pPAC-β-galactosidase expression vector (1 μg per 60-mm dish) was included in all transfections and its expression was used for normalization of the luciferase activity. This graph represents data from one of three individual experiments that showed similar results.
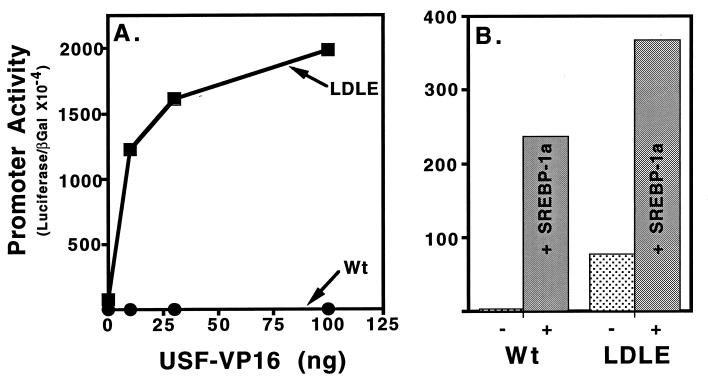
We predicted that other bHLH proteins might bind and activate the E-box in the mutant LDL receptor promoter. Therefore, we evaluated this directly by a transfection assay in mammalian cells where the wild-type and mutant LDL receptor promoters were transfected into HepG2 cells along with constructs that express a hybrid between the bHLH protein USF2 and the potent activation domain from the herpes simplex virus type I VP16 protein (31). USF2 is a closely related bHLH protein to the SREBPs and the E-box present in the mutant LDL receptor was predicted to bind USF2 with high affinity (26). Transfection of increasing amounts of the USF2-VP16 expression resulted in an efficient dose-dependent activation of the E-box containing LDL receptor mutant promoter but the wild-type LDL receptor promoter was unresponsive (Fig. 3A ▪). In contrast, both promoters were activated by cotransfection of a vector that expresses the mature truncated version of SREBP-1a (Fig. 3B).
Figure 3.
Activation of the wild-type and mutant LDL receptor promoter by USF2-VP16. (A). HepG2 cells were transfected with increasing amounts of plasmid cytomegalovirus-USF2-VP16 expression vector (abscissa) and wild-type (Wt; •) or mutant (LDLE; ▪) LDL receptor promoter luciferase reporter plasmids (5 μg per 60-mm dish) along with 5 μg per 60-mm dish of cytomegalovirus-β-galactosidase plasmid as indicated. (B) Companion dishes were transfected with the same reporter plasmids alone (−) or along with the SREBP-1a expression vector (+) as indicated (30 ng per 60-mm dish). Transfections were performed and analyzed as described in Materials and Methods and in the legend to Fig. 2.
The experiments of Fig. 3 were performed in cells cultured in medium containing lipoprotein derived cholesterol where the concentration of endogenous nuclear SREBPs would be minimal and the basal level of the wild-type LDL receptor promoter is quite low. Interestingly the basal level of promoter activity for the E-box containing variant was several-fold higher than for the wild-type LDL receptor (Fig. 3B), which is consistent with activation of this mutant promoter by endogenous bHLH proteins that are targeted to the nucleus independently of the cholesterol requirement of the cell.
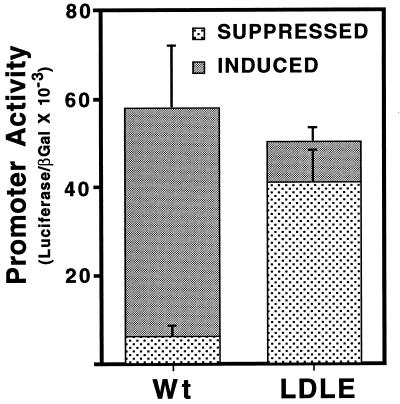
The experiments presented so far indicate that SREBP-1a can bind to and activate the wild-type and mutant LDL receptor promoters similarly but that a classic E-box binding protein specifically activated the mutant LDL receptor containing the E-box substitution but had no stimulatory effect on the wild-type SRE containing promoter. Next, we evaluated the ability of both promoters to be regulated by exogenously added sterols. Dishes of CV-1 cells were transfected with either the wild-type or E-box mutant promoter and were subsequently cultured in medium containing either lipoprotein-depleted serum (Induced) or the identical medium supplemented with a mixture of cholesterol and 25-hydroxycholesterol (Suppressed). Consistent with previous studies (16, 18) the wild-type promoter was expressed several-fold higher when the transfected cells were cultured in the absence of sterols (Induced vs. Suppressed, Fig. 4). However, the mutant promoter was expressed at a relatively high level regardless of the sterol content of the culture media (LDLE). These results are consistent with a loss of sterol regulation of the mutant promoter due to its promiscuous activation by constitutively expressed E-box binding proteins such as USF2.
Figure 4.
Evaluation of sterol regulation for the wild-type and mutant LDL receptor promoters. The wild-type (Wt) or mutant (LDLE) LDL receptor promoter fused to the luciferase gene were transfected into CV-1 cells (5 μg per 60-mm dish) and 18 hr after transfection dishes were fed either medium containing lipoprotein-depleted serum (Induced) or medium containing lipoprotein-depleted serum plus 10 μg/ml of cholesterol and 1 μg/ml of 25-hydroxycholesterol (Suppressed). The cells were cultured for an additional 24 hr before harvest. The samples were processed as described in Materials and Methods. Each value represents the average of two independent experiments each performed in duplicate.
DISCUSSION
The current experiments were designed to address the regulatory significance of the special DNA recognition properties of the SREBPs. As members of the bHLH family they bind to the palindromic E-box element but they also bind to the direct repeat SRE as well as to several variant SREs. The key observation that stimulated the current investigation was that although SREBPs bind to both types of sites, sterol regulation in all promoters analyzed carefully to date is mediated through SREBP binding to SRE elements and not the classic E-box palindrome. There are several related bHLH proteins that are expressed and constitutively targeted to the nucleus that could bind and activate transcription through recognizing E-box sites independent of the sterol level of the cell. Thus, we hypothesized that SRE recognition provided a mechanism to allow selectivity and specificity in the sterol regulatory response because SREBPs would be present in the nucleus and bind to SREs only when the cell needed more cholesterol.
To test this hypothesis we converted the direct repeat SRE of the human LDL receptor promoter into a classic E-box and then evaluated the mutant promoter for its ability to support SREBP-mediated transcription and sterol regulation. The results clearly demonstrated that SREBPs are capable of activating transcription through E-box sites with the same requirement for coregulatory factors as seen with the wild-type LDL receptor promoter (Figs. 2 and 3). Additionally, the E-box containing mutant promoter (but not the wild-type LDL receptor promoter) was activated by a related bHLH protein that binds only to E-box sites (Fig. 3). Most importantly, the results of Fig. 4 show that the native LDL receptor promoter was efficiently regulated by sterols in transfected cells, whereas the E-box variant was not. A key observation was that the activity of this mutant promoter under conditions of high sterol levels was nearly equal to that under low sterol culture conditions. This is consistent with activation of the E-box containing promoter by endogenous constitutively expressed bHLH binding proteins independent of the cholesterol level of the cell. In contrast, the activity of the wild-type LDL receptor promoter was relatively low when sterols were added to the cultured cells because under these conditions the concentration of nuclear SREBPs is very low and the endogenous E-box recognizing proteins are incapable of binding efficiently to the SRE to mediate transcriptional activation.
As mentioned above, the ability of SREBPs to bind palindromic E-boxes and direct repeat SREs is largely caused by a tyrosine in the basic region of the bHLH motif (26). The only other bHLH proteins that contain this tyrosine residue are from the yeast Saccharomyces cerevisiae. One of them, TYE7, was identified as an activator of Ty1-mediated gene expression (37). Independently, two alleles of the same gene were identified as second-site suppressors of a null mutation of the GCR1 gene. In this second study the gene was named SGC1 (38). Because GCR1 is an activator of genes required for glucose utilization, the TYE7/SGC1 gene is likely to be involved in the regulation of carbohydrate metabolizing genes. Thus, it is tempting to suggest that TYE7/SGC1 and the SREBPs evolved from a common ancestor that was a global nutritional regulator. The two additional yeast genes predicted to contain the special tyrosine were identified from the yeast genome sequencing project and the functional roles for these additional proteins is uncertain. Although the yeast genes contain the special tyrosine residue in their basic domains, they totally lack the C-terminal region that anchors the precursor SREBPs to membranes and confers sterol-regulated nuclear accumulation to the mammalian proteins.
We have expressed the TYE7/SGC1 protein in E. coli and have shown the purified protein binds both direct repeat SREs and palindromic E-boxes (unpublished data). Thus, the DNA binding specificity appears to be similar to the SREBPs and the significance of the dual recognition for this yeast regulator remains to be established.
There are other distantly related bHLH proteins that have divergent basic DNA binding domains and they fail to bind specifically to the conserved E-box motif. These include the enhancer of split protein family of Drosophila melanogaster that appear to recognize the so called “N-box” (39) as well as the mammalian aryl hydrocarbon receptor that binds the xenobiotic response element, which is located in the regulatory region of genes that are required for the metabolism of xenobiotics (40). Interestingly, a domain located just N-terminal to the classic basic domain is required for efficient DNA recognition of some variant SREs by the SREBPs (30) and a similarly positioned domain is required for recognition of xenobiotic response element by the aryl hydrocarbon receptor (40). Thus, there are other examples where the amino acid residues in and around the basic domain of bHLH proteins have evolved to recognize divergent DNA sites but they have retained the same fundamental protein structural motif. This suggests that the bHLH structure provides an efficient scaffold for properly orienting amino acid side chains for specific DNA recognition, which is a conclusion supported by a comprehensive alanine scanning mutagenesis scheme that was performed on the bHLH domain (41).
In a previous report from our laboratory (9) we demonstrated that activation of the FAS promoter by endogenously produced nuclear SREBP that occurs when cells are cultured in the absence of an exogenous supply of cholesterol was mediated by two distinct adjacent SREBP binding sites one between −77 and −63 and the other between −61 and −54. Importantly, each of these SREBP binding sites contains half of an E-box element that is located between −65 and −62. We identified the two SREBP binding sites by DNase I footprinting experiments with wild-type and mutant promoters that separately disrupted each predicted SREBP binding site. In fact, some of the mutations that inhibited SREBP binding to one site or the other left the E-box element unaltered. Importantly, we also demonstrated that sterol regulation of the FAS promoter by endogenously produced SREBPs was significantly reduced by mutations that altered either of the SREBP sites. Again, some of the mutants that were defective for activation by endogenously produced SREBPs left the E-box element intact.
In contrast, the authors of a recent report (42) suggested that insulin regulation of the FAS promoter was mediated by SREBP-1c action through the E-box element located at position −65 to −62. This conclusion was based on two observations: (i) a mutation that simultaneously disrupted both halves of the E-box decreased activation by high levels of ectopically expressed SREBP-1c and (ii) expression of high levels of a mutant version of SREBP-1c that substituted an arginine in place of the critical tyrosine residue of the basic region and restricts SREBP binding only to E-boxes still activated the FAS promoter.
Because both halves of the E-box reside within the SREBP binding sites identified in our previous study conclusions based on a single mutation that simultaneously disrupts the E-box and both SREBP sites must be interpreted cautiously. Additionally, the data from our current studies also suggests that overexpression of SREBPs can clearly activate gene expression through E-box sites (Fig. 3). Therefore, firm conclusions for activation by the wild-type SREBPs based on results for activation by high levels of expression of a mutant protein designed to bind solely to E-box sites are also limited. When taken together all of the available data are consistent with activation of the FAS promoter by endogenously expressed wild-type SREBPs through the tandem SREBP sites.
It is possible that this FAS E-box is the target for insulin regulation by a nonsterol-regulated E-box binding protein as has been suggested recently by Wang and Sul (43). Because the biosynthesis of fatty acids and cholesterol need to be independently as well as coordinately regulated, the activation of FAS through the E-box by more specific regulators of fatty acid metabolism would provide such a mechanism.
Because our present experiments demonstrate (Fig. 3B) that ectopic expression of SREBPs can activate the LDL receptor promoter when its SRE element is converted into an E-box, it is likely that endogenously produced SREBPs do activate promoters with E-box sites. However, the E-boxes must be correctly positioned close to sites for the required coregulatory factors such as Sp1 and nuclear factor-Y, which is a requirement for SREBPs to efficiently activate gene expression (16–19, 44). However, nonsterol-regulated endogenous E-box proteins could also activate such promoters so the difference in promoter activity before and after sterol depletion would be minimal and the effective regulation would be modest at best.
The coevolution mechanism described here for achieving specificity in gene regulation by SREBPs is unusual. Other bHLH proteins achieve specificity in various ways: the expression of MyoD, a muscle specific differentiation factor, is tissue restricted and its activity is negatively regulated through heterodimerization with another protein—called Id—that contains the HLH motif but lacks the basic domain (45). In other cases the DNA binding and activation potential of bHLH proteins is determined by their intrinsic ability to function as homo or heterodimers with other related proteins. For example, c-Myc only binds DNA efficiently as a heterodimer with Max and thus activation by c-Myc is dependent on the coexpression of Max or a related protein (46).
The coevolution of DNA binding site and the associated binding protein can also help to explain the target gene specificity of the nuclear receptor family of proteins. In this case, specific amino acids in and around the conserved zinc fingers of the DNA binding domain in the individual receptor proteins dictate slightly different target gene specificity (47).
Nonetheless, the current experiments address a fundamental question in the evolution of metabolic regulatory mechanisms and establish the existence of a mode of coevolution of regulatory protein and DNA recognition site. The SREBPs have diverged from the classic bHLH DNA binding family through the acquisition of a specific tyrosine residue in their DNA binding domain. This allows efficient recognition of direct repeat SRE elements that have evolved to confer cholesterol-regulated transcription to key target genes. The coevolution of the DNA binding protein along with its recognition site has ensured maximal specificity and sensitivity in the sterol regulatory response.
Acknowledgments
We thank Joseph Goldstein and Michael Brown for comments on the manuscript and Howard Towle for the gift of the USF-VP16 plasmid. T.F.O. is an Established Investigator of the American Heart Association. This work was supported in part by grants from the National Institutes of Health (HL48044) and the American Heart Association (93001330 and 96008190).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: SRE, sterol regulatory element; SREBP, SRE binding proteins; bHLH, basic helix–loop–helix proteins; E-box, a palindromic DNA sequence (5′-CANNTG-3′); FAS, fatty acid synthase; LDL, low density lipoprotein; LDLE, low density lipoprotein E-box; USF, upstream stimulatory factor.
References
- 1.Goldstein J, Brown M. Nature (London) 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 2.Osborne T F. Crit Rev Eukaryotic Gene Expression. 1995;5:317–335. doi: 10.1615/critreveukargeneexpr.v5.i3-4.50. [DOI] [PubMed] [Google Scholar]
- 3.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 4.Smith J R, Osborne T F, Brown M S, Goldstein J L, Gil G. J Biol Chem. 1988;263:18480–18487. [PubMed] [Google Scholar]
- 5.Osborne T F. J Biol Chem. 1991;266:13947–13951. [PubMed] [Google Scholar]
- 6.Ericsson J, Jackson S M, Lee B C, Edwards P A. Proc Natl Acad Sci USA. 1996;93:945–950. doi: 10.1073/pnas.93.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ericsson J, Jackson S M, Kim J B, Spiegelman B M, Edwards P A. J Biol Chem. 1997;272:7298–7305. doi: 10.1074/jbc.272.11.7298. [DOI] [PubMed] [Google Scholar]
- 8.Guan G, Dai P-H, Osborne T F, Kim J B, Shechter I. J Biol Chem. 1997;272:10295–10302. doi: 10.1074/jbc.272.15.10295. [DOI] [PubMed] [Google Scholar]
- 9.Magaña M M, Osborne T F. J Biol Chem. 1996;271:32689–32694. doi: 10.1074/jbc.271.51.32689. [DOI] [PubMed] [Google Scholar]
- 10.Magaña M M, Lin S S, Dooley K A, Osborne T F. J Lipid Res. 1997;38:1630–1638. [PubMed] [Google Scholar]
- 11.Yokoyama C, Wang X, Briggs M R, Admon A, Wu J, Hua X, Goldstein J L, Brown M S. Cell. 1993;75:185–197. [PubMed] [Google Scholar]
- 12.Hua X, Yokoyama C, Wu J, Briggs M R, Brown M S, Goldstein J L, Wang X. Proc Natl Acad Sci USA. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Sato R, Brown M S, Hua X, Goldstein J L. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 14.Hua X, Sakai J, Ho Y K, Goldstein J L, Brown M S. J Biol Chem. 1995;270:29422–29427. doi: 10.1074/jbc.270.49.29422. [DOI] [PubMed] [Google Scholar]
- 15.Sakai J, Duncan E A, Rawson R B, Hua X, Brown M S, Goldstein J L. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 16.Sudhof T, Van der Westhuyzen D, Goldstein J, Brown M, Russell D. J Biol Chem. 1987;262:10773–10779. [PubMed] [Google Scholar]
- 17.Briggs M R, Yokoyama C, Wang X, Brown M S, Goldstein J L. J Biol Chem. 1993;268:14490–14496. [PubMed] [Google Scholar]
- 18.Sanchez H B, Yieh L, Osborne T F. J Biol Chem. 1995;270:1161–1169. doi: 10.1074/jbc.270.3.1161. [DOI] [PubMed] [Google Scholar]
- 19.Jackson S M, Ericsson J, Osborne T F, Edwards P A. J Biol Chem. 1995;270:21445–21448. doi: 10.1074/jbc.270.37.21445. [DOI] [PubMed] [Google Scholar]
- 20.Dooley K A, Millinder S, Osborne T F. J Biol Chem. 1998;273:1349–1356. doi: 10.1074/jbc.273.3.1349. [DOI] [PubMed] [Google Scholar]
- 21.Bennett M K, Lopez J M, Sanchez H B, Osborne T F. J Biol Chem. 1995;270:25578–25583. doi: 10.1074/jbc.270.43.25578. [DOI] [PubMed] [Google Scholar]
- 22.Lopez J M, Bennett M K, Sanchez H B, Rosenfeld J M, Osborne T F. Proc Natl Acad Sci USA. 1996;93:1049–1053. doi: 10.1073/pnas.93.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimano H, Horton J D, Hammer R E, Shimomura I, Brown M S, Goldstein J L. J Clin Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murre C, Bain G, van Dijk M A, Engel I, Furnari B A, Massari M E, Matthews J R, Quong M W, Rivera R R, Stuiver M H. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 25.Blackwell T K, Huang J, Ma A, Kretzner L, Alt F W, Eisenman R N, Weintraub H. Mol Cell Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J B, Spotts G D, Halvorsen Y-D, Shih H-M, Ellenberger T, Towle H C, Spiegelman B M. Mol Cell Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sudhof T, Russell D, Brown M, Goldstein J. Cell. 1987;48:1061–1069. doi: 10.1016/0092-8674(87)90713-6. [DOI] [PubMed] [Google Scholar]
- 28.Smith J R, Osborne T F, Goldstein J L, Brown M S. J Biol Chem. 1990;265:2306–2310. [PubMed] [Google Scholar]
- 29.Smith J R. Ph.D. thesis. Dallas: Univ. of Texas Southwestern Medical School; 1990. [Google Scholar]
- 30.Millinder-Vallett S, Sanchez H B, Rosenfeld J M, Osborne T F. J Biol Chem. 1996;271:12247–12253. doi: 10.1074/jbc.271.21.12247. [DOI] [PubMed] [Google Scholar]
- 31.Kaytor E N, Shih H, Towle H C. J Biol Chem. 1997;272:7525–7531. doi: 10.1074/jbc.272.11.7525. [DOI] [PubMed] [Google Scholar]
- 32.Athanikar J N, Sanchez H B, Osborne T F. Mol Cell Biol. 1997;17:5193–5200. doi: 10.1128/mcb.17.9.5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldstein J, Basu S, Brown M. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 35.Tontonoz P, Kim J B, Graves R A, Spiegelman B M. Mol Cell Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yieh L, Sanchez H B, Osborne T F. Proc Natl Acad Sci USA. 1995;92:6102–6106. doi: 10.1073/pnas.92.13.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lohning C, Ciriacy M. Yeast. 1994;10:1329–1339. doi: 10.1002/yea.320101010. [DOI] [PubMed] [Google Scholar]
- 38.Nishi K, Park C S, Pepper A E, Eichinger G, Innis M, Holland M J. Mol Cell Biol. 1995;15:2646–2653. doi: 10.1128/mcb.15.5.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oellers N, Dehio M, Knust E. Mol Gen Genet. 1994;244:465–473. doi: 10.1007/BF00583897. [DOI] [PubMed] [Google Scholar]
- 40.Fukunaga B N, Hankinson O. J Biol Chem. 1996;271:3743–3749. doi: 10.1074/jbc.271.7.3743. [DOI] [PubMed] [Google Scholar]
- 41.Fisher D E, Parent L A, Sharp P A. Cell. 1993;72:467–476. doi: 10.1016/0092-8674(93)90122-7. [DOI] [PubMed] [Google Scholar]
- 42.Kim J B, Sarraf P, Wright M, Yao K M, Mueller E, Solanes G, Lowell B B, Speigelman B M. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang D, Sul H S. J Biol Chem. 1997;272:26367–26374. doi: 10.1074/jbc.272.42.26367. [DOI] [PubMed] [Google Scholar]
- 44.Ericsson J, Jackson S M, Edwards P A. J Biol Chem. 1996;271:24359–24364. doi: 10.1074/jbc.271.40.24359. [DOI] [PubMed] [Google Scholar]
- 45.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 46.Blackwood E M, Eisenman R N. Science. 1991;251:1211–1216. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 47.Umesono K, Evans R M. Cell. 1989;57:1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]