Abstract
The nonselective human corticotropin-releasing factor receptor 1 (hCRF-R1) and the ligand-selective Xenopus CRF-R1 (xCRF-R1) were compared. To understand the interactions of sauvagine and ovine CRF, both high-affinity ligands for hCRF-R1 but surprisingly weak ligands for xCRF-R1, chimeric receptors of hCRF-R1 and xCRF-R1 followed by double or multiple point mutations were constructed. Binding studies and cAMP assays demonstrated that the N-terminal domain exhibited the complete ligand selectivity of xCRF-R1. The important region was mapped between amino acids 70 and 89; replacement of amino acids Arg76, Asn81, Gly83, Leu88, and Ala89 in hCRF-R1 with the corresponding amino acids of xCRF-R1 (Gln76, Gly81, Val83, His88, and Leu89) resulted in a receptor that had ∼30-fold higher affinity for human/rat CRF than for sauvagine. Mutagenesis of these amino acids in xCRF-R1 to the human sequence completely abolished the ligand selectivity of xCRF-R1. Mutagenesis of amino acids 88 and 89 in hCRF-R1 or xCRF-R1 had only a minor (∼2.5-fold) effect on the ligand selectivity of the mutant receptor. Substitution of Arg76, Asn81, and Gly83 in hCRF-R1 with the corresponding sequence of xCRF-R1 (Gln76, Gly81, and Val83) resulted in a receptor with ∼11-fold higher affinity for human/rat CRF compared with ovine CRF or sauvagine. When only two of these three amino acids were mutated, no effect on the ligand selectivity was observed. On the basis of these data, it is suggested that amino acids 70–89 of CRF-R1 are important for the ligand binding site.
Corticotropin-releasing factor (CRF), a 41-amino acid peptide (1) originally isolated by its ability to regulate the secretion of adrenocorticotropic hormone in the anterior pituitary (2), is the main mediator of the stress response (3, 4). Two CRF receptor (CRF-R) subtypes, both belonging to the superfamily of G protein-coupled receptors (GPCR), have been identified in vertebrate species. cDNAs encoding CRF-Rs type 1 (CRF-R1) and type 2 (CRF-R2) have been isolated from various species including human (5–7), rat (8–10), mouse (6, 11–13), and chicken (14).
The ligand-binding domains of GPCRs that bind small molecules such as catecholamines (15, 16) or acetylcholine (17) are well characterized and were mapped to the transmembrane domains. Additionally, for GPCRs that are activated by small peptides such as vasotocin (18), substance P (19–21), or somatostatin-14 (22), the ligand binding pocket has been mapped to the transmembrane regions (TMs) and the extracellular loops between TM2 and TM7. For receptors that are activated by larger peptides such as secretin or vasoactive intestinal polypeptide, it has been demonstrated that the first extracellular domain (EC) is involved in the high-affinity ligand binding (23).
Mammalian CRF-R1 in contrast to mammalian CRF-R2 has been reported to be nonselective for CRF from different species and the structurally related CRF analogs urocortin (24, 25), urotensin I (26), and sauvagine (27). All of these peptides were bound with the same affinity and were able to stimulate intracellular cAMP accumulation with similar potency (7, 10, 11, 14, 24, 25). Recently, we have isolated CRF-R1 from the amphibian species Xenopus laevis (28) that was highly ligand-selective, despite its high degree of identity (>80%) on amino acid level to human CRF-R1 (hCRF-R1). The CRF analogs human/rat CRF (h/rCRF), Xenopus CRF (xCRF; ref. 29), urotensin I, and urocortin were bound by xCRF-R1 with higher affinity than the structurally related analogs ovine CRF (oCRF) and sauvagine. The regions responsible for the remarkable ligand selectivity of xCRF-R1 may play an important role in ligand binding and activation of the CRF-R. Therefore, it is of high importance to identify these different residues to understand the interaction of the CRF ligands with their receptors.
To map the domain involved in the ligand selectivity of xCRF-R1, we constructed a series of chimeric receptors in which various parts of the N termini of both receptors were replaced by the corresponding domains of their counterparts. The ligand-selective residues of xCRF-R1 were determined by double or multiple amino acid mutations.
MATERIALS AND METHODS
Materials, Peptides, and Reagents.
All cell culture reagents were purchased from GIBCO/BRL, and aprotinin was obtained from Boehringer Mannheim. CRF analogs were synthesized by solid-phase methods as described (30). The purity was greater than 98%.
Construction of Chimeric Receptors, Mutagenesis, and Nomenclature.
Chimeric receptors were constructed from hCRF-R1 or xCRF-R1 cDNA, inserted in the pcDNA3 vector (Invitrogen) at restriction enzyme sites generated by PCR with Pfu DNA polymerase (Stratagene). For the generation of multiple amino acid substitutions the Exsite kit (Stratagene) was used, whereas double amino acid exchanges were accomplished with the QuickChange kit (Stratagene). Sequences from PCR or synthetic oligonucleotides were verified by DNA sequencing with an Applied Biosystems model 373 DNA sequencer; the GCG software package was used for analysis. In chimeric receptors such as h1(1–123)x1(124–415), the origin of the N-terminal residues are named first, whereas the origin of the remaining receptor parts are listed last. In mutated receptors such as h1x1(H88,L89), first the receptor in which the mutagenesis was performed is indicated and then the name of the receptor from which the mutagenized residues were selected is indicated; the residues that replaced the original amino acids (in this case amino acids 88 and 89 of xCRF-R1) are in parentheses in the single-letter amino acid code.
Radioreceptor Binding Assays.
Isolation of membranes from permanently transfected HEK 293 cells expressing xCRF-R1, hCRF-R1, chimeric, or mutant receptors and Scatchard analysis were performed with 50–100 μg of protein as described (28). The Bmax values for all chimeric and mutant receptors when tested with the three CRF analogs did not differ significantly from each other (2.4 ± 0.4 pmol/mg of membrane protein).
cAMP Assay.
HEK 293 cells stably transfected with cDNA coding for xCRF-R1, hCRF-R1, chimeric, or mutant receptors were plated at 105 cells per well into 24-well dishes. Stimulation for 30 min (37°C, 5% CO2) with peptides and intracellular cAMP assays were performed as described (28). Statistical analysis was performed by two-way ANOVA; significant differences between groups were determined by post hoc analysis using the Dunnett test.
RESULTS
Ligand Selectivity of hCRF-R1, xCRF-R1, and Six Chimeric Receptors.
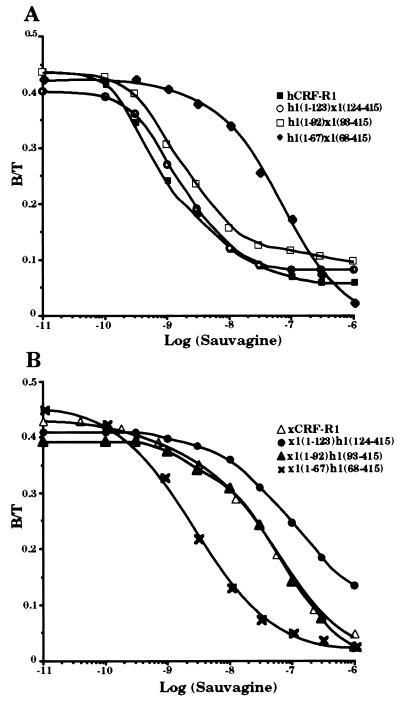
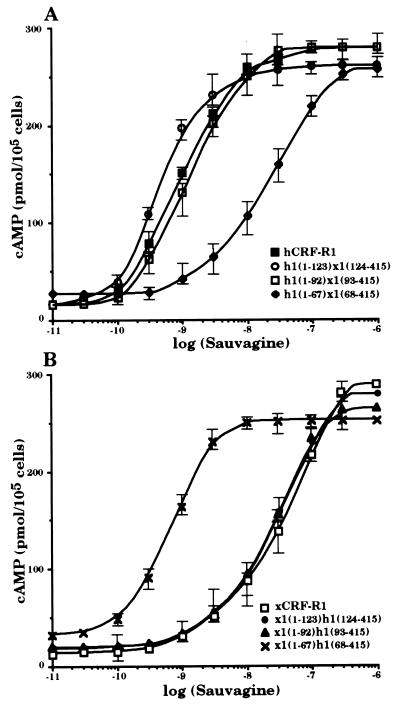
cDNAs coding for six chimeric receptors, h1(1–123)x1(124–415), h1(1–92)x1(93–415), h1(1–67)x1(68–415), x1(1–123)h1(124–415), x1(1–92)h1(93–415), and x1(1–67)h1(68–415), were constructed so that the cDNA sequences coding for the N-terminal parts of either hCRF-R1 or xCRF-R1 were decreased (Fig. 1). These constructs were stably transfected into HEK 293 cells. The receptors produced h1(1–123)x1(124–415) and h1(1–92)x1(93–415), like hCRF-R1, bound the tested CRF analogs h/rCRF, oCRF, and sauvagine with similar affinity (Fig. 2 and Table 1). The chimeric receptors x1(1–123)h1(124–415), x1(1–92)h1(93–415), and xCRF-R1 displayed ligand selectivities that did not differ significantly from one another. Thus, the entire ligand selectivity of xCRF-R1 was located in its N-terminal domain. However, when only the first 67 N-terminal amino acids of either xCRF-R1 or hCRF-R1 were replaced with the corresponding amino acids of their counterpart, the ligand selectivity of the modified receptors was not affected (Fig. 2 and Table 1). h/rCRF (Kd = 2.5 ± 1.0 nM) was bound by h1(1–67)x1(68–415) with ∼15-fold higher affinity than oCRF (Kd = 30.2 ± 2.6 nM) or sauvagine (Kd = 36.9 ± 4.6 nM). On the other hand, the affinities of x1(1–67)h1(68–415) for h/rCRF, oCRF, and sauvagine, respectively, did not differ significantly from each other. The ability of the various receptor chimeras to stimulate cAMP production in HEK 293 cells stably transfected with the corresponding cDNAs was tested with the same ligands used in the binding experiments. Ligand preferences similar to those in the binding experiments were observed (Fig. 3 and Table 2). Sauvagine was slightly (∼2-fold) more potent to stimulate cAMP production in HEK 293 cells stably transfected with hCRF-R1 or h1(1–123)x1(124–415) than h/rCRF or oCRF. Stimulation of cAMP accumulation in HEK 293 cells stably transfected with h1(1–92)x1(93–415) or x1(1–67)h1(68–415) resulted in similar dose–response curves for the tested CRF analogs that were without significant differences. The potency of h/rCRF to stimulate cAMP production in cells stably transfected with the chimeric receptors x1(1–123)h1(124–415), x1(1–92)h1(93–415), and h1(1–67)x1(68–415) was significantly higher than the potency of oCRF or sauvagine (Table 2). h/rCRF was 25- to 30-fold more potent than sauvagine. Thus, it was concluded that the ligand selective domain of xCRF-R1 resided between amino acids 68 and 92.
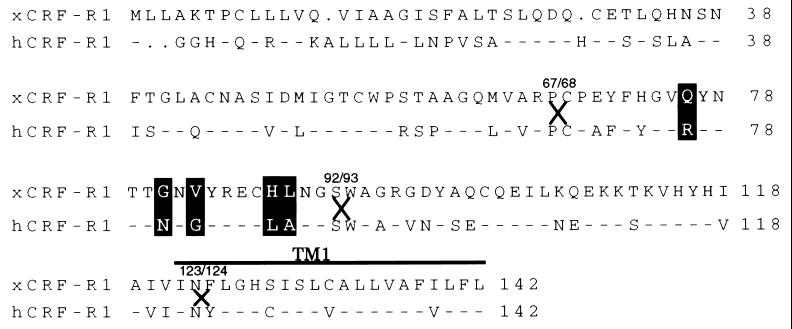
Figure 1.
Schematic representation and sequence comparison of EC1 and TM1 domains of xCRF-R1 and hCRF-R1. The conserved amino acids within EC1 and TM1 are presented as dashes in the hCRF-R1 sequence. The residues that have been mutated are highlighted, and the junctions of chimeric receptors (residues 67/68, 92/93, and 123/124) are indicated by X.
Figure 2.
Binding of sauvagine to membranes of HEK 293 cells transfected with hCRF-R1, h1(1–123)x1(124–415), h1(1–92)x1(93–415), and h1(1–67)x1(68–415) (A) or xCRF-R1, x1(1–123)h1(124–415), x1(1–92)h1(93–415), and x1(1–67)h1(68–415) (B). Competitive binding was performed with 125I-labeled h/rCRF and increasing concentrations (0.01 nM to 1 μM) of unlabeled sauvagine. Data represent duplicates from one representative experiment repeated at least three times.
Table 1.
Binding properties of hCRF-R1, xCRF-RI, and six chimeric receptors
| Receptor | h/rCRF Kd, nM | oCRF Kd, nM | Sauvagine Kd, nM |
|---|---|---|---|
| hCRF-R1a | 2.2 ± 1.2 | 2.6 ± 1.1 | 2.1 ± 1.1 |
| h1(1–123)x1(124–415) | 1.8 ± 1.2 | 1.9 ± 0.3 | 1.6 ± 1.0 |
| h1(1–92)x1(93–415) | 1.6 ± 0.7 | 2.9 ± 1.5 | 2.2 ± 1.2 |
| h1(1–67)x1(68–415) | 2.5 ± 1.0 | 30.2 ± 2.6* | 36.9 ± 4.6* |
| x1(1–67)h1(68–415) | 2.4 ± 1.1 | 3.4 ± 2.0 | 2.1 ± 1.3 |
| x1(1–92)h1(93–415) | 3.0 ± 1.4 | 31.2 ± 4.6* | 43.9 ± 4.2*§ |
| x1(1–123)h1(124–415) | 4.4 ± 2.1 | 36.3 ± 3.9† | 61.4 ± 9.2*§ |
| xCRF-R1 | 3.1 ± 1.4 | 31.7 ± 2.6† | 51.4 ± 6.6*‡ |
Data are the mean ± SEM of three binding studies. Statistically significant differences:
P < 0.0001 vs. h/rCRF.
P < 0.001 vs. h/rCRF
P < 0.001 vs. oCRF.
P < 0.01 vs. oCRF.
From ref. 28.
Figure 3.
Stimulation of intracellular cAMP accumulation of HEK 293 cells stably transfected with hCRF-R1, h1(1–123)x1(124–415), h1(1–92)x1(93–415), and h1(1–67)x1(68–415) (A) or xCRF-R1, x1(1–123)h1(124–415), x1(1–92)h1(93–415), and x1(1–67)h1(68–415) (B) by sauvagine. Cells were incubated with increasing concentrations of sauvagine for 30 min at 37°C, and cAMP was determined. The results are representatives of three stimulations.
Table 2.
Stimulation of cAMP accumulation by different CRF-like peptides in HEK 293 cells expressing hCRF-R1, xCRF-R1, or six different chimeric receptor cDNAs
| Receptor | h/rCRF EC50, nM | oCRF EC50, nM | Sauvagine EC50, nM |
|---|---|---|---|
| hCRF-R1a | 1.6 ± 0.2 | 1.8 ± 0.2 | 0.9 ± 0.1†¶ |
| h1(1–123)x1(124–415) | 0.8 ± 0.2 | 1.2 ± 0.1 | 0.4 ± 0.1‡∥ |
| h1(1–92)x1(93–415) | 0.9 ± 0.1 | 1.3 ± 0.2 | 1.1 ± 0.2 |
| h1(1–67)x1(68–415) | 0.7 ± 0.2 | 4.1 ± 0.3* | 20.9 ± 0.5*§ |
| x1(1–67)h1(68–415) | 0.8 ± 0.2 | 1.4 ± 0.3 | 1.0 ± 0.2 |
| x1(1–92)h1(93–415) | 0.7 ± 0.1 | 4.9 ± 0.7* | 20.9 ± 0.6*§ |
| x1(1–123)h1(124–415) | 0.8 ± 0.2 | 6.5 ± 0.9* | 26.1 ± 0.7*§ |
| xCRF-R1 | 1.1 ± 0.1 | 7.1 ± 0.8* | 39.7 ± 0.8*§ |
Data are the mean ± SEM of three stimulations. Statistically significant differences:
P < 0.0001 vs. h/rCRF.
P < 0.01 vs. h/rCRF
P < 0.05 vs. h/rCRF.
P < 0.0001 vs. oCRF.
P < 0.001 vs. oCRF.
∥ P < 0.01 vs. oCRF.
From ref. 28.
Mapping of the Amino Acids Involved in the Ligand Selectivity of xCRF-R1 by Point Mutations.
To further map the amino acids for ligand selectivity of xCRF-R1, double and multiple point mutations were created between residues 68 and 92.
Substitution of five amino acids in xCRF-R1 with the corresponding residues of hCRF-R1, Arg76, Asn81, Gly83, Leu88, and Ala89, generating x1h1(R76,N81,G83,L88,A89), almost completely abolished the ligand selectivity of the mutant receptor (Table 3). h/rCRF (Kd = 3.8 ± 1.2 nM), oCRF (Kd = 4.4 ± 2.3 nM), and sauvagine (Kd = 4.5 ± 2.7 nM) displaced [Tyr0]h/rCRF with affinities that did not differ significantly from one another. Substitution of five residues in hCRF-R1 with the corresponding amino acids of xCRF-R1, Gln76, Gly81, Val83, His88, and Leu89, generating h1x1(Q76,G81,V83,H88,L89), resulted in a ligand-selective receptor mutant. h/rCRF (Kd = 2.6 ± 1.3 nM) was bound with more than 13-fold higher affinity than oCRF (Kd = 31.9 ± 4.1 nM) or sauvagine (Kd = 34.9 ± 4.7 nM). Binding studies with h1x1(H88,L89) revealed a slightly preferential binding of h/rCRF (Kd = 1.8 ± 0.8 nM) compared with oCRF (Kd = 5.2 ± 1.4 nM) and sauvagine (Kd = 4.5 ± 2.1 nM), respectively. However, these differences were not statistically significant (Table 3). Replacement of residues 88 and 89 in xCRF-R1 produced a small but significant effect in the binding properties of the resulting receptor mutant x1h1(L88,A89). Although h/rCRF was bound with higher affinity (Kd = 1.3 ± 0.3 nM) than oCRF (Kd = 12.6 ± 3.5 nM) or sauvagine (Kd = 11.2 ± 2.4 nM), the effect was smaller than with xCRF-R1.
Table 3.
Binding properties of different mutants of hCRF-R1 or xCRF-R1
| Receptor | h/rCRF Kd, nM | oCRF Kd, nM | Sauvagine Kd, nM |
|---|---|---|---|
| h1x1(Q76,G81,V83,H88,L89) | 2.6 ± 1.3 | 31.9 ± 4.1* | 34.9 ± 4.7* |
| h1x1(H88,L89) | 1.8 ± 0.8 | 5.2 ± 1.4 | 4.5 ± 2.1 |
| h1x1(Q76,G81,V83) | 2.5 ± 1.5 | 28.4 ± 2.0* | 25.6 ± 4.8* |
| h1x1(Q76,G81) | 1.4 ± 0.9 | 2.4 ± 0.4 | 1.5 ± 0.5 |
| h1x1(G81,V83) | 3.5 ± 1.9 | 4.4 ± 2.4 | 3.9 ± 1.5 |
| x1h1(R76,N81,G83,L88,A89) | 3.8 ± 1.2 | 4.4 ± 2.3 | 4.5 ± 2.7 |
| x1h1(L88,A89) | 1.3 ± 0.3 | 12.6 ± 3.5† | 11.2 ± 2.4† |
Data are the mean ± SEM of three binding studies. Statistically significant differences:
P < 0.001 vs. h/rCRF.
P < 0.01 vs. h/rCRF.
Replacement of amino acids 76, 81, and 83 in hCRF-R1 with the corresponding residues of xCRF-R1 had a strong effect on the ligand selectivity of the mutant receptor molecule. h1x1(Q76,G81,V83) bound h/rCRF (Kd = 2.5 ± 1.5 nM) with more than 10-fold higher affinity than oCRF (Kd = 28.4 ± 2.0 nM) and sauvagine (Kd = 25.6 ± 4.8 nM). In contrast to h1x1(Q76,G81,V83), both h1x1(Q76,G81) and h1x1(G81,V83) mutants, respectively, bound the tested CRF analogs with similar selectivity (Table 3).
When stimulation of cAMP production in HEK 293 cells stably transfected with cDNAs coding for the various mutant receptors was performed (Table 4), it was observed that h/rCRF (EC50 = 0.7 ± 0.2 nM) was 30-fold more potent in h1x1(Q76,G81,V83,H88,L89)-transfected HEK 293 cells than sauvagine (EC50 = 20.4 ± 0.8 nM). x1h1(R76,N81,G83,L88,A89) expressed in HEK 293 cells did not discriminate significantly among h/rCRF, oCRF, or sauvagine, used to stimulate cAMP production. HEK 293 cells transfected with cDNA coding for h1x1(H88,L89) responded to h/rCRF stimulation (EC50 = 1.0 ± 0.1 nM) at slightly but significantly lower peptide concentrations than to oCRF (EC50 = 2.8 ± 0.2 nM) or sauvagine (EC50 = 2.6 ± 0.3 nM). HEK 293 cells stably transfected with cDNA coding for x1h1(L88,A89) displayed a higher ligand selectivity when stimulated to produce cAMP than HEK 293 cells transfected with cDNA coding for h1x1(H88,L89). h/rCRF (EC50 = 1.2 ± 0.2 nM) was ∼9-fold more potent than oCRF (EC50 = 10.3 ± 0.5 nM) or sauvagine (EC50 = 10.2 ± 0.9 nM), but the difference between the potencies of h/rCRF and sauvagine was ∼3-fold smaller than in HEK 293 cells transfected with cDNA coding for h1x1(Q76,G81,V83,H88,L89) (Table 4).
Table 4.
Stimulation of cAMP accumulation of different CRF-like peptides in HEK 293 cells expressing different mutants of hCRF-R1 or xCRF-R1 cDNAs
| Receptor | h/rCRF EC50, nM | oCRF EC50, nM | Sauvagine EC50, nM |
|---|---|---|---|
| h1x1(Q76,G81,V83,H88,L89) | 0.7 ± 0.2 | 4.9 ± 0.4* | 20.4 ± 0.8*† |
| h1x1(H88,L89) | 1.0 ± 0.1 | 2.8 ± 0.2* | 2.6 ± 0.3* |
| h1x1(Q76,G81,V83) | 0.9 ± 0.2 | 8.3 ± 1.0* | 10.1 ± 0.7* |
| h1x1(Q76,G81) | 1.1 ± 0.2 | 1.4 ± 0.3 | 1.3 ± 0.3 |
| h1x1(G81,V83) | 2.2 ± 0.2 | 2.7 ± 0.3 | 2.4 ± 0.3 |
| x1h1(R76,N81,G83,L88,A89) | 1.0 ± 0.2 | 1.4 ± 0.2 | 1.2 ± 0.1 |
| x1h1(L88,A89) | 1.2 ± 0.2 | 10.3 ± 0.5* | 10.2 ± 0.9* |
Data are the mean ± SEM of three stimulations. Statistically significant differences:
P < 0.0001 vs. h/rCRF.
P < 0.0001 vs. oCRF.
As observed with the binding assays, mutation of residues 76, 81, and 83, respectively, to generate h1x1(Q76,G81,V83), resulted in a strong discrimination among the CRF analogs used in this study. Stimulation of HEK 293 cells stably transfected with cDNA coding for this receptor mutant with h/rCRF resulted in cAMP production at low agonist concentrations (EC50 = 0.9 ± 0.2 nM), whereas oCRF (EC50 = 8.3 ± 1.0 nM) and sauvagine (EC50 = 10.1 ± 0.7 nM) activated the receptor at significantly higher peptide concentrations (Table 4). No significant difference in the potency of the tested CRF analogs to stimulate cAMP production in HEK 293 cells stably transfected with cDNA coding for either h1x1(Q76,G81) or for h1x1(G81,V83) was observed.
DISCUSSION
Chimeric receptors have been successfully used to map regions involved in the agonist/antagonist binding of several GPCRs (18, 21, 23, 31, 32).
We used this approach followed by generation of point mutations, in the current study, to identify regions that are involved in the remarkable ligand selectivity of xCRF-R1. This receptor weakly bound sauvagine, a potent ligand for all CRF-Rs known to date. Binding studies and cAMP assays were used to monitor the affinities and potencies of various chimeric and mutant receptors. Because xCRF-R1 had a 10- to 20-fold higher binding affinity for h/rCRF compared with oCRF and sauvagine, respectively (28), we tested these three CRF analogs to identify the ligand-selective domain of xCRF-R1.
Both xCRF-R1 and hCRF-R1 were found to be highly homologous on amino acid level with an overall identity of ∼80%. The highest sequence homology was found in the intracellular loops (96% identity) and the transmembrane regions (89% identity), whereas the ECs were only 66% identical (28). The lowest degree of identity in ECs was observed in the N-terminal domain EC1, which was conserved by only 57% between xCRF-R1 and hCRF-R1. The other ECs showed a much higher degree of sequence conservation with 71% identity in EC2, 91% identity in EC3, and 88% identity in EC4. Thus, it was expected that EC1 was strongly involved in the ligand selectivity of xCRF-R1.
Binding assays and cAMP studies supported this hypothesis. x1(1–123)h1(124–415) displayed the same ligand selectivity as xCRF-R1. Thus, we concluded that the ligand selectivity of xCRF-R1 resided in its N-terminal domain. Sequence comparison of the EC1 domain in hCRF-R1 and xCRF-R1 showed that most of the divergence between both receptors was found in the first 67 residues, which were only 51% identical, whereas the middle part of EC1 (residues 68–92) and the part close to TM1 (residues 93–121) were 68% and 62% identical, respectively.
Surprisingly, as shown in Results, neither amino acid residues 1–67 nor residues 93–121, the most diverging regions in EC1, influenced the ligand selectivity of xCRF-R1. The chimeric receptor x1(1–92)h1(93–415) retained almost completely the ligand selectivity of xCRF-R1. Further shortening of the EC1 domain (amino acids 1–67) had no influence on the ligand selectivity of the constructed receptors chimeras. Thus, we concluded that the ligand-selective region of xCRF-R1 was located between residues 68 and 92.
To further support this finding, point mutations were introduced into hCRF-R1 and xCRF-R1. Eight of 25 amino acids were found to diverge between xCRF-R1 and hCRF-R1 within this domain. Further sequence comparison revealed that Glu70 and Tyr71 of xCRF-R1 were conserved in chicken CRF-R1 (cCRF-R1, ref. 14) and mammalian CRF-R2 (7, 10–13), two high-affinity sauvagine receptors. No sequence conservation was observed for His74, Gln76, Gly81, Val83, His88, and Leu89 of xCRF-R1 in comparison to other CRF-Rs. It is important to note that hCRF-R1 and cCRF-R1 contain identical amino acids in this domain (Tyr74, Arg76, Asn81, Gly83, Leu88, and Ala89; refs. 5, 6, and 14). Thus, it was expected that the ligand selectivity of xCRF-R1 was determined by these residues. With the exception of His74, the other residues were taken into consideration when point mutations were introduced into hCRF-R1 and xCRF-R1, respectively. As shown in Results, mutation of amino acids 76, 81, 83, 88, and 89 of hCRF-R1 to the xCRF-R1 sequence created a CRF-R with ligand selectivity that was nearly identical to that of xCRF-R1. Conversely, replacement of the residues described above in xCRF-R1 with the corresponding amino acids of hCRF-R1 almost completely abolished the ligand discrimination of the resulting mutant receptor. When residues Leu88 and Ala89 in hCRF-R1 were replaced by His88 and Leu89 of xCRF-R1, only a minor effect on the ligand selectivity was observed (see Results). Consistently, replacement of these amino acids in xCRF-R1 with the corresponding amino acids of hCRF-R1 resulted in a ∼3-fold drop of the ligand selectivity. Single point mutations of residues 88 or 89 had no effect on the selectivity of the mutant receptors (data not shown). It was concluded that both residues contribute to the ligand-selective site of xCRF-R1.
In contrast to the minor effects of amino acids 88 and 89, mutagenesis of residues 76, 81, and 83, respectively, to the xCRF-R1 sequence (Gln76, Gly81, and Val83) created a ligand-selective receptor. Thus, h1x1(Q76,G81,V83) displayed a ∼11-fold higher affinity for h/rCRF than for oCRF or sauvagine. However, the ligand selectivity of this receptor mutant was significantly weaker than the one of h1x1(Q76G81V83H88L89) or xCRF-R1. The involvement of all three residues in the ligand selectivity of xCRF-R1 was demonstrated when only two of these amino acids (positions 76 and 81 or position 81 and 83) were mutated in hCRF-R1. Both receptor mutants bound the tested CRF analogs unselectively with high affinity. Thus, it was concluded that five amino acids in the EC1 domain of xCRF-R1 are responsible for the ligand selectivity of this receptor.
At this point, it is important to note that oCRF was a more potent stimulator of cAMP production than sauvagine in HEK 293 cells stably transfected with cDNAs coding for several chimeric receptors and the receptor mutant h1x1(Q76G81V83H88L89), although both peptides displayed similarly low affinity in the binding experiments. The differences observed in our study might be because of a longer half-life of intact oCRF as observed in earlier studies (33). Furthermore, the activation of a small population of receptor molecules can result in a maximal cAMP response in a transfection system used herein. Therefore, small differences in the affinities of the peptides can result in larger differences in their potencies to activate the second messenger cascade. Therefore, we believe that the differences in the binding affinities of the various CRF analogs are a stronger indication for their potencies in a natural system with fewer receptors. In addition cAMP stimulations were carried out at 37°C, whereas the binding assays were performed at room temperature. Because Xenopus laevis is normally maintained at room temperature, it cannot be completely excluded that at 37°C ligand receptor interactions may have been enhanced by receptor folding. However, it is very unlikely that the data presented in this study are caused by an artifact because the differences in the affinities of xCRF-R1 to the ligands shown in Results and in our previous publication (28) were observed in the binding experiments and with small variations in the stimulation of cAMP production. Furthermore, the mutation of five amino acids in hCRF-R1 to the corresponding residues of xCRF-R1 created a ligand-selective hCRF-R that is expected to be fully active at 37°C.
The data presented herein contrast with the findings of Liaw et al. (34), who mapped the ligand selectivity of hCRF-R2, a receptor binding sauvagine with higher affinity than h/rCRF. The amino acids involved in the ligand selectivity of hCRF-R2 were mapped to three regions of its EC2 and EC3 domains (region 1, Ile172-Asp173-His174 in EC2; region 2, His185 in EC2; region 3, Asp262-Leu263-Val264 in EC3; ref. 31). Replacement of the original residues in hCRF-R1 with the corresponding amino acids of regions 1–3 in hCRF-R2 created a receptor mutant displaying the ligand selectivity of hCRF-R2. Interestingly, the ligand-selective domains of xCRF-R1 (28) and hCRF-R2 (7, 34) resided in different regions of the receptor molecules. It is important to note that hCRF-R1 and xCRF-R1 are identical on amino acid level in the equivalents of the ligand-selective regions 2 and 3 in hCRF-R2. Only in the equivalent of region 1 (Thr175-Leu176-Ser177-His178 in hCRF-R1 or Thr175-Met176-Ser177-Pro178 in xCRF-R1) were two amino acid differences between both type 1 receptors identified, but the two different residues were not conserved in hCRF-R2. Furthermore, Liaw et al. (34) did not show that the replacement of the ligand-selective domains of hCRF-R2 with the corresponding regions of hCRF-R1 created a nonselective CRF-R. Therefore, the involvement of the above mentioned regions in the ligand recognition of CRF-R1 remains unclear at this time. Because mammalian CRF-R1 is a nonselective receptor (7, 10, 11, 14, 24, 25, 28), it seems likely that modifications in the EC1 domain might have resulted in the loss of the ligand selectivity of CRF-R1 during evolution. The importance of the entire EC1 domain in high-affinity ligand binding of amphibian and mammalian CRF-R1 is further supported by the finding that chimeric receptors in which the EC1 domain of rat growth hormone-releasing factor receptor was substituted with the EC1 domain of rat CRF-R1 still bound CRF analogs with high affinity (35). Similar data were obtained for other peptide receptors including the receptors for pituitary adenylate cyclase activating peptide, vasoactive intestinal polypeptide, or secretin (23, 36). Therefore it seems likely that the amino acids responsible for the ligand selectivity of xCRF-R1 represent a critical part of the binding pocket of CRF-R1.
Because it can be expected that different parts of the ECs in CRF-Rs are involved in the high-affinity ligand binding, it is conceivable that the region we have mapped is an important part of the CRF binding pocket. Interestingly, the identity of residues 68–92 of EC1 in all CRF-Rs known to date is 64%, the highest sequence identity score within the EC1 domain. This high degree of sequence conservation within the CRF-R family (5–11, 13, 14, 28) contrasts with a relatively low degree (28–32%) of sequence identity of CRF-R(68–92) with other closely related receptors such as rat growth-hormone releasing factor receptor (31), secretin receptor (32), or vasoactive intestinal polypeptide receptor (37, 38). Thus, the importance of this region for the substrate specificity of CRF-R1 has been demonstrated.
In conclusion, we have identified a region within the EC1 domain of xCRF-R1 that transduces the remarkable ligand selectivity of this CRF-R and gives insights in the receptor–ligand interactions of CRF-Rs.
Acknowledgments
We thank Dr. Ulrich Teichert and Ulrike Schulz for synthesis of oligonucleotides. Dr. Andreas Rühmann and Ines Bonk are gratefully acknowledged for synthesis of the peptides used in this investigation. In addition, we thank Konstanze Dietrich and Monika Palchaudhuri for their help with some of the membrane preparations and cAMP stimulations. We also thank Almuth Burgdorf for her help in the processing of the manuscript.
ABBREVIATIONS
- CRF-R
CRF receptor
- oCRF
ovine CRF
- h/rCRF
human/rat CRF
- xCRF
Xenopus CRF
- GPCR
G protein-coupled receptor
- EC
extracellular domain
- TM
transmembrane domain
References
- 1.Spiess J, Rivier J, Rivier C, Vale W. Proc Natl Acad Sci USA. 1981;78:6517–6521. doi: 10.1073/pnas.78.10.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vale W, Spiess J, Rivier C, Rivier J. Science. 1981;213:1394–1397. doi: 10.1126/science.6267699. [DOI] [PubMed] [Google Scholar]
- 3.Dunn A J, Berridge C W. Brain Res Rev. 1990;15:71–100. doi: 10.1016/0165-0173(90)90012-d. [DOI] [PubMed] [Google Scholar]
- 4.Owens M J, Nemeroff C B. Pharmacol Rev. 1991;43:425–473. [PubMed] [Google Scholar]
- 5.Chen R, Lewis K A, Perrin M H, Vale W W. Proc Natl Acad Sci USA. 1993;90:8967–8971. doi: 10.1073/pnas.90.19.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vita N, Laurent P, Lefort S, Chalon P, Lelias J M, Kaghad M, Le Fur G, Caput D, Ferrara P. FEBS Lett. 1993;335:1–5. doi: 10.1016/0014-5793(93)80427-v. [DOI] [PubMed] [Google Scholar]
- 7.Liaw C W, Lovenberg T W, Barry G, Oltersdorf T, Grigoriadis D E, De Souza E B. Endocrinology. 1996;137:72–77. doi: 10.1210/endo.137.1.8536644. [DOI] [PubMed] [Google Scholar]
- 8.Perrin M H, Donaldson C J, Chen R, Lewis K A, Vale W W. Endocrinology. 1993;133:3058–3061. doi: 10.1210/endo.133.6.8243338. [DOI] [PubMed] [Google Scholar]
- 9.Chang C P, Pearse R V, II, O’Connel S, Rosenfeld M G. Neuron. 1993;11:1187–1195. doi: 10.1016/0896-6273(93)90230-o. [DOI] [PubMed] [Google Scholar]
- 10.Lovenberg T W, Liaw C W, Grigoriadis D E, Clevenger W, Chalmers D T, De Souza E B, Oltersdorf T. Proc Natl Acad Sci USA. 1995;92:836–840. doi: 10.1073/pnas.92.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kishimoto T, Pearse R V, II, Lin C R, Rosenfeld M G. Proc Natl Acad Sci USA. 1995;92:1108–1112. doi: 10.1073/pnas.92.4.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perrin M, Donaldson C, Chen R, Blunt A, Berggren T, Bilezikjian L, Sawchenko P, Vale W. Proc Natl Acad Sci USA. 1995;92:2969–2973. doi: 10.1073/pnas.92.7.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenzel P, Kesterson R, Yeung W, Cone R D, Rittenberg M B, Stenzel-Poore M P. Mol Endocrinol. 1995;9:637–645. doi: 10.1210/mend.9.5.7565810. [DOI] [PubMed] [Google Scholar]
- 14.Yu J, Xie L Y, Abou-Samra A B. Endocrinology. 1996;137:192–197. doi: 10.1210/endo.137.1.8536612. [DOI] [PubMed] [Google Scholar]
- 15.Dixon R A, Sigal I S, Rands E, Register R B, Candelore M R, Blake A D, Strader C D. Nature (London) 1987;326:73–77. doi: 10.1038/326073a0. [DOI] [PubMed] [Google Scholar]
- 16.Strader C D, Sigal I S, Dixon R A F. FASEB J. 1989;3:1825–1832. doi: 10.1096/fasebj.3.7.2541037. [DOI] [PubMed] [Google Scholar]
- 17.Wheatley M, Hulme E C, Birdsall N J M, Curtis C A M, Eveleigh P, Pedder E K, Poyner D. Trends Pharmacol Sci Suppl. 1988;9:19–24. [PubMed] [Google Scholar]
- 18.Hausmann H, Richters A, Kreienkamp H J, Meyerhof W, Mattes H, Lederis K, Zwiers H, Richter D. Proc Natl Acad Sci USA. 1996;93:6907–6912. doi: 10.1073/pnas.93.14.6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fong T M, Huang R R C, Strader C D. J Biol Chem. 1992;267:25664–25667. [PubMed] [Google Scholar]
- 20.Huang R R C, Yu H, Strader C D, Fong T M. Biochemistry. 1994;33:3007–3013. doi: 10.1021/bi00176a033. [DOI] [PubMed] [Google Scholar]
- 21.Huang R R C, Yu H, Strader C D, Fong T M. Mol Pharmacol. 1994;45:690–695. [PubMed] [Google Scholar]
- 22.Nehring R, Meyerhof W, Richter D. DNA Cell Biol. 1995;14:939–944. doi: 10.1089/dna.1995.14.939. [DOI] [PubMed] [Google Scholar]
- 23.Gourlet P, Vilardaga J P, de Neef P, Waelbroeck M, Vandermeers A, Robberecht P. Peptides. 1996;17:825–829. doi: 10.1016/0196-9781(96)00107-6. [DOI] [PubMed] [Google Scholar]
- 24.Vaughan J, Donaldson C, Bittencourt J, Perrin M H, Lewis K, Sutton S, Chan R, Turnbull A V, Lovejoy D, Rivier C, et al. Nature (London) 1995;378:287–292. doi: 10.1038/378287a0. [DOI] [PubMed] [Google Scholar]
- 25.Donaldson C, Sutton S, Perrin M H, Corrigan A Z, Lewis K A, Rivier J, Vaughan J M, Vale W W. Endocrinology. 1996;137:2167–2170. doi: 10.1210/endo.137.5.8612563. [DOI] [PubMed] [Google Scholar]
- 26.Lederis K, Letter A, McMaster D, Moore G. Science. 1982;218:162–164. doi: 10.1126/science.6981844. [DOI] [PubMed] [Google Scholar]
- 27.Montecucchi P C, Henschen A. Int J Peptide Protein Res. 1981;18:113–120. doi: 10.1111/j.1399-3011.1981.tb02047.x. [DOI] [PubMed] [Google Scholar]
- 28.Dautzenberg F M, Dietrich K, Palchaudhuri M R, Spiess J. J Neurochem. 1997;69:1640–1645. doi: 10.1046/j.1471-4159.1997.69041640.x. [DOI] [PubMed] [Google Scholar]
- 29.Stenzel-Poore M P, Heldwein K A, Stenzel P, Vale W W. Mol Endocrinol. 1992;6:1716–1724. doi: 10.1210/mend.6.10.1448118. [DOI] [PubMed] [Google Scholar]
- 30.Rühmann A, Köpke A K E, Dautzenberg F M, Spiess J. Proc Natl Acad Sci USA. 1996;93:10609–10613. doi: 10.1073/pnas.93.20.10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayo K E. Mol Endocrinol. 1992;6:1734–1744. doi: 10.1210/mend.6.10.1333056. [DOI] [PubMed] [Google Scholar]
- 32.Ishihara T, Nakamura S, Kaziro Y, Takahashi T, Takahashi K, Nagata S. EMBO J. 1991;10:1635–1641. doi: 10.1002/j.1460-2075.1991.tb07686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orth D N, De Bold C R, De Cherney G S, Jackson R V, Sheldon W R, Jr, Nicholson W E, Uderman H, Alexander A N, Island D P, Rivier J, Vale W W. Fed Proc. 1985;44:197–202. [PubMed] [Google Scholar]
- 34.Liaw C W, Grigoriadis D E, Lovenberg T W, De Souza E B, Maki R A. Mol Endocrinol. 1997;11:980–985. doi: 10.1210/mend.11.7.9946. [DOI] [PubMed] [Google Scholar]
- 35.Perrin M H, Sutton S, Bain D, Berggren W T, Vale W W. Endocrinology. 1998;139:566–570. doi: 10.1210/endo.139.2.5757. [DOI] [PubMed] [Google Scholar]
- 36.Hashimoto H, Ogawa N, Hagihara N, Yamamoto K, Imanishi K, Nogi H, Nishino A, Fujita T, Matsuda T, Nagata S, Baba A. Mol Pharmacol. 1997;52:128–135. doi: 10.1124/mol.52.1.128. [DOI] [PubMed] [Google Scholar]
- 37.Svoboda M, Tastenoy M, Van Rampelbergh J, Goossens J F, De Neef P, Waelbroeck M, Robberecht P. Biochem Biophys Res Commun. 1994;205:1617–1624. doi: 10.1006/bbrc.1994.2852. [DOI] [PubMed] [Google Scholar]
- 38.Ishihara T, Shigemoto R, Mori K, Takahashi K, Nagata S. Neuron. 1992;8:811–819. doi: 10.1016/0896-6273(92)90101-i. [DOI] [PubMed] [Google Scholar]