Abstract
To investigate the mechanism of SWI/SNF action, we have analyzed the pathway by which SWI/SNF stimulates formation of transcription factor-bound nucleosome core complexes. We report here that the SWI/SNF complex binds directly to nucleosome cores and uses the energy of ATP hydrolysis to disrupt histone/DNA interactions, altering the preferred path of DNA bending around the histone octamer. This disruption occurs without dissociating the DNA from the surface of the histone octamer. ATP-dependent disruption of nucleosomal DNA by SWI/SNF generates an altered nucleosome core conformation that can persist for an extended period after detachment of the SWI/SNF complex. This disrupted conformation retains an enhanced affinity for the transcription factor GAL4-AH. Thus, ATP-dependent nucleosome core disruption and enhanced binding of the transcription factor can be temporally separated. These results indicate that SWI/SNF can act transiently in the remodeling of chromatin structure, even before interactions of transcription factors.
The SWI/SNF complex is a 2-MDa multiprotein complex that contains the products of several genes originally identified in yeast by defects in mating type switching (SWI) and/or sucrose fermentation (SNF). The SWI/SNF complex is required for the transcription of a subset of yeast genes and also for the function of several heterologous transcription activators in yeast (reviewed in refs. 1 and 2). Homologues of SWI/SNF components also have been identified in other organisms (3–5). In mammals, two close homologues of the SWI2 gene are brg1 and hbrm (6–8). Biochemical fractionations indicated that there are at least two distinct SWI/SNF complexes in mammalian cells (9, 10), and the subunit composition of these SWI/SNF complexes varies in different cell lines (9). In Drosophila, brahma (brm) is the closest homologue of yeast SWI2 and is likely to encode a component of the Drosophila SWI/SNF complex (11, 12). brm is required for embryogenesis and regulates the expression of many homeotic genes (11). In addition, two more distant SWI2 relatives, the Drosophila ISWI gene (13) and the yeast STH1 gene (14), have been found to encode part of smaller complexes, termed NURF, CHRAC, ACF (ISWI), and RSC (STH1), that are distinct from SWI/SNF but have in vitro activities somewhat similar to SWI/SNF (15–19) (see below).
A connection between the SWI/SNF complex and chromatin structure was first evidenced by the fact that swi/snf mutants can be suppressed by mutations in histone genes and other nonhistone chromosomal proteins (20–23). Moreover, SWI/SNF has been shown to alter the in vivo chromatin structure at the SWI-dependent SUC2 promoter in a manner independent of transcription (22, 23). In vitro analysis of the yeast and mammalian SWI/SNF complexes supports a role in remodeling chromatin structures. Both yeast and mammalian complexes are able to disrupt DNase I digestion patterns of rotationally phased nucleosomal DNA, and stimulate nucleosome core binding by GAL4 derivatives, TBP, Sp1, NF-κB, and USF in a manner dependent on ATP hydrolysis (9, 10, 24–26).
Low-resolution nuclease digestion studies of nucleosome arrays in vitro illustrate both persistent and reversible effects of SWI/SNF action. The yeast SWI/SNF complex was found to create a transcription factor-dependent DNase I-hypersensitive site within an array of nucleosomes in vitro that persists after removal of the SWI/SNF complex (27). This permanent change in the structure of the nucleosome array was the manifestation of histone eviction caused by a combination of SWI/SNF action and the binding of five GAL4 dimers, which further destabilizes the nucleosomes. Nucleosomes within an array that are not bound by GAL4 dimers appear to lose defined nucleosomal boundaries (27) and acquire increased restriction endonuclease accessibility at SWI/SNF action (28). However, these effects on nucleosome arrays appear to require continued SWI/SNF binding and ATP hydrolysis as these properties are reversed at detachment of the SWI/SNF complex and/or removal of ATP (27, 28).
ATP hydrolysis is not continuously required for the mammalian SWI/SNF complex to stimulate the binding of transcription factors to nucleosomes. Using high-resolution DNase mapping, Imbalzano et al. (29) have shown that once the SWI/SNF complex has disrupted nucleosome core structure, ATP can be depleted without loss of the disrupted state of the nucleosome cores. The remaining SWI/SNF-bound nucleosome cores retain an enhanced affinity for GAL4-AH after ATP removal (29). These results suggest a function of the SWI/SNF complex in nucleosome core disruption that does not require continued ATP hydrolysis but instead may be maintained by continued SWI/SNF binding and/or by a stable alteration of nucleosome core structure. Because our earlier studies detected reversible changes in low-resolution DNase I and restriction endonuclease digestion of nucleosome arrays mediated by the SWI/SNF complex (above), we tested the persistence of SWI/SNF effects on nucleosome core structure and activator binding after detachment of the SWI/SNF complex.
We report here that disruption of the path of DNA bending around the histone octamer by the SWI/SNF complex persists after detachment of the SWI/SNF complex. Moreover, the disrupted conformation of the nucleosome core can persist for several hours after SWI/SNF detachment and retains an enhanced affinity for GAL4-AH. These data indicate that nucleosome core disruption and stimulation of activator binding by SWI/SNF can be temporally separated. Thus, transient action of SWI/SNF may remodel chromatin structures before the stable interactions of transcription activators.
MATERIALS AND METHODS
DNA Probes.
The end-labeled 176-bp 5S DNA probe containing a nucleosome positioning sequence was prepared as described (27). For the experiments in Fig. 5, probes were produced from pBend5S(172) containing a single copy of 172-bp 5S DNA cloned in the EcoRV site of pBend1. PCR was performed by using EcoRI–HindIII primers from pBR322. The 195-bp product then was digested with XhoI and PvuII. The probe was body-labeled during the PCR or end-labeled at the XhoI end by the Klenow fragment of DNA polymerase. Production of the 154-bp probe with a single GAL4 site 32 bp from the end using pBend401G1 was done as described (24).
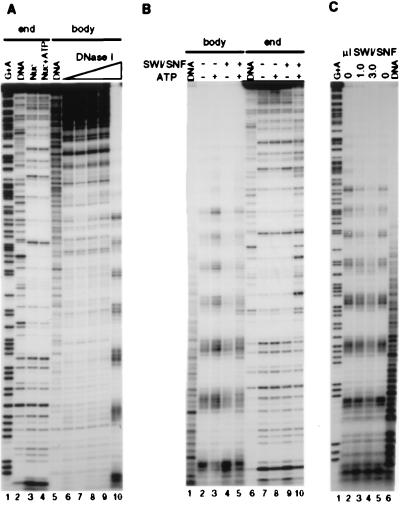
Figure 5.
SWI/SNF alters the path of DNA bending around the histone octamer (A) The same core 5S DNA sequence as in Fig. 1 was end-labeled (lanes 2–4) or body-labeled (lanes 5–10), reconstituted into nucleosome cores, and digested with DNase I. Mock-reconstituted DNA is shown in lanes 2 and 5. 25 ng (12.5 nM) of total nucleosomes (probe plus donor oligonucleosomes) was incubated as before, followed by digestion with 0.05 unit (lanes 1, 5), 0.5 unit (lanes 3, 4), 1 unit (lane 8), 2 units (lane 9), or 10 units (lane 10) of DNase I; 1 mM Mg-ATP is present in lanes 4 and 7. (B) Body-labeled (lanes 1–5) and end-labeled (lanes 6–10) reconstituted probes were incubated in the presence or the absence of 2.5 nM SWI/SNF and/or ATP followed by digestion with 10 units (lanes 2–5) or 0.5 unit (lanes 7–10) of DNase I. Mock reconstitutions were digested with 1 unit (lane 1) and 0.05 unit (lane 6) of DNase I. (C) Reactions were performed as in B with the body-labeled reconstituted probe (using 10 units of DNase I) except that ATP is present in all lanes and 7.5 nM SWI/SNF is used in lane 4. Note the single 10-nucleotide digestion product kept in the gel.
Protein Purification and Nucleosome Core Reconstitutions.
HeLa H1-depleted oligonucleosomes were prepared as described (30). Yeast SWI/SNF complex and GAL4-AH transcription factor were purified essentially as described (24). Nucleosome core reconstitutions on labeled probes were performed by the octamer transfer method (30). Naked DNA controls were prepared by adding the probe DNA at the end of the transfer protocol, except in Fig. 1, where the naked DNA lanes contained phenol-extracted HeLa oligonucleosomes.
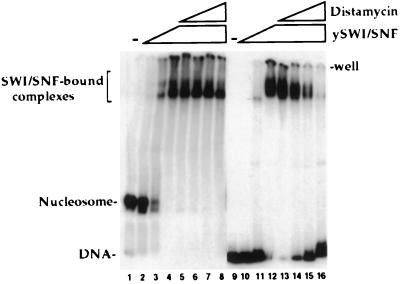
Figure 1.
SWI/SNF complex binding to nucleosome cores or naked DNA. Purified SWI/SNF complex was incubated with a 176-bp 5S RNA gene probe either as reconstituted nucleosome cores (lanes 1–8) or as naked DNA (lanes 9–16). SWI/SNF was present at 0 nM in lanes 1 and 9, 3 nM in lanes 2 and 10, 10 nM in lanes 3 and 11, and 30 nM in lanes 4–8 and 12–16. Distamycin A was present during SWI/SNF binding in lanes 5 and 13 (0.1 μM), lanes 6 and 14 (1 μM), lanes 7 and 15 (10 μM), and lanes 8 and 16 (100 μM). All lanes contain 9 nM total nucleosomal or naked DNA (mainly HeLa unlabeled oligonucleosomes or DNA) and 1 mM Mg-ATP. Samples were run on 4% acrylamide gel (80:1, acrylamide/bis) in a Tris/glycine/EDTA buffer system.
Preparation and Analysis of Binding Reactions.
For gel shift assays 10-μl binding reactions were prepared in 20 mM Hepes-NaOH pH 7.5/50 mM NaCl/3 mM MgCl2/1 μM ZnCl2/0.1% Nonidet P-40/5% glycerol/2 mM DTT/0.2 mM phenylmethylsulfonyl fluoride/0.2 mg/ml BSA. Unlabeled donor nucleosomes (12.5 ng) were present with 0.25 ng of probe DNA. GAL4-AH, 1 mM Mg-ATP, and SWI/SNF were added when indicated. The reactions were incubated for 30 min at 30°C and loaded directly on a 4% acrylamide (29:1)/0.5× TBE gel, except in Fig. 1 where 4% acrylamide(80:1)/1× TGE gel was used. For DNase I footprinting assays the binding reactions were doubled and treated as described (24) (0.5 unit of DNase I for nucleosomes and 0.05 unit for mock reconstituted DNA). For the binding reactions involving apyrase treatment, 0.025 (gel shifts) or 0.050 (DNase I) unit of apyrase (Sigma) was added at the indicated time, and the reaction was incubated for at least 20 min at 30°C (up to 90 min after SWI/SNF action). SWI/SNF competition was performed by adding 20-fold mass excess of H1-depleted HeLa oligonucleosomes at the end of the binding reaction followed by a 30-min incubation at 30°C; except for Fig. 4 where time-course competition was performed by using 20-fold mass excess of both oligonucleosomes and calf thymus DNA at 37°C. In Fig. 1, distamycin A was added to the binding reaction and incubated 5 min on ice before SWI/SNF addition.
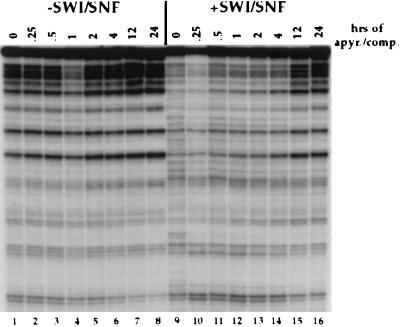
Figure 4.
Perturbation of nucleosome core structure by SWI/SNF is stable for several hours after its detachment but eventually reverts back. Time-course experiment on the stability of the altered nucleosomal state induced by SWI/SNF. 5S nucleosome cores were incubated with 15 nM SWI/SNF complex for 30 min at 30°C in the presence of 1 mM ATP. Apyrase and 20-fold excess of both unlabeled HeLa oligonucleosomes and calf thymus DNA then were added. Reactions were put at 37°C for the indicated time before DNase I addition.
RESULTS
SWI/SNF Binding to Nucleosome Cores.
The SWI/SNF complex interacts with nucleosome cores independently of whether the nucleosome core is bound by transcription activators. To better understand SWI/SNF interactions with nucleosome cores, we have begun to analyze the requirements for this binding. Quinn and colleagues (31) have found that the SWI/SNF complex binds to naked DNA with a Kd in the range of 1–9 nM. This binding was relatively sequence-independent and resembled that of high mobility group (HMG)-box-containing proteins. The binding of SWI/SNF complex to naked DNA appears to occur through minor groove interactions and the complex is displaced from DNA by the minor groove-binding reagent distamycin A (31). As shown in the low bis acrylamide gel in Fig. 1, the SWI/SNF complex also binds directly to nucleosome cores with an affinity slightly higher than that for naked DNA (2- to 3-fold). Binding of the complex to DNA (31) and nucleosomes (Fig. 1) is independent of ATP. However, the binding of SWI/SNF to nucleosome cores differs from binding to naked DNA in that the complex is not readily displaced from nucleosome cores by distamycin A. This distamycin A-resistant binding suggests that the binding of SWI/SNF to nucleosome cores includes additional or distinct interactions that further stabilize the binding relative to binding naked DNA. The only difference between the nucleosome core and DNA probes in Fig. 1 is the presence of the histone octamer that indicates that the distamycin A resistance of SWI/SNF binding to nucleosome cores is mediated via the histone proteins (e.g., either structurally or by protein–protein interactions).
Detachment of the SWI/SNF Complex Leaves a Persistently Altered Nucleosome Core.
The ability to assay the interaction of SWI/SNF with nucleosome cores (as shown in Fig. 1) allowed us to investigate the steps in its function that required the physical association of the SWI/SNF complex with the nucleosome core substrates. Whereas the SWI/SNF complex binds to nucleosome cores with nanomolar affinity, its binding is largely sequence-independent. Thus, SWI/SNF can be detached from bound nucleosome cores by competition with an excess of cold oligonucleosomes. In Fig. 2A, labeled 5S nucleosome cores were incubated with sufficient SWI/SNF to achieve complete binding (lane 2). Note that in contrast to Fig. 1, the standard gel shift system used in Fig. 2 does not allow SWI/SNF-nucleosome core complexes to substantially enter the gel. In Fig. 2A lanes 3 and 4 the initial binding reaction was followed by a 30-min incubation with a 20-fold excess of cold H1-depleted HeLa oligonucleosomes. This incubation efficiently detached SWI/SNF from the 5S nucleosome cores (lane 4), which then migrated equivalent to the unbound nucleosome cores in lane 3. The reappearance of the nucleosome core after SWI/SNF competition indicates that histones are still present in the nucleosome cores after ATP-dependent SWI/SNF action, as suggested previously (24).
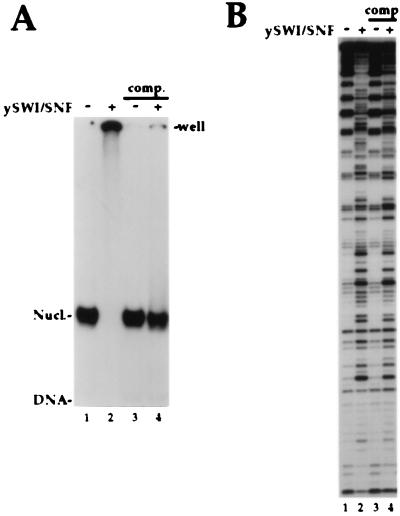
Figure 2.
Dissociation of the SWI/SNF complex leaves a persistently altered nucleosome core. (A) A 5S DNA probe as in Fig. 1 was reconstituted into nucleosome cores. SWI/SNF binding was analyzed by standard gel shift assay [4% acrylamide (29/1)/0.5× TBE]. Each reaction contains 12.5 nM total nucleosomes and 1 mM Mg-ATP, whereas 30 nM SWI/SNF was added in lanes 2 and 4. In lanes 3 and 4, after an initial 30-min incubation, 20-fold excess of cold HeLa oligonucleosomes were added followed by another 30-min incubation at 30°C before loading on the gel. (B) Binding reactions similar to A were performed and analyzed by DNase I footprint assay. 15 nM of SWI/SNF was added in lanes 2 and 4. Competitors and 2-fold more DNase I were used in lanes 3 and 4. Near stochiometric ratio of SWI to nucleosomes were used here and in the following experiments to insure almost complete binding of the nucleosomes in the initial reaction.
To test whether the disruption of nucleosome cores by SWI/SNF would persist after its detachment, DNase digestions were performed by using reactions similar to those shown in Fig. 2A. The DNase digestion patterns of these nucleosome cores are shown in Fig. 2B. The unbound nucleosome cores in lane 1 presents the typical DNase I pattern of cutting for rotationally phased nucleosomal 5S DNA (cuts detected every 10–11 bp). The rotational phasing of the 5S nucleosome cores was unaffected by the addition of the competitors (Fig. 2B, lane 3). By contrast, in presence of SWI/SNF and ATP the pattern of cutting was dramatically changed, reflecting the perturbation of histone-DNA contacts by SWI/SNF (loss of the 10-bp-repeating pattern; lane 2). Surprisingly, the SWI/SNF-perturbed digestion pattern of 5S nucleosome cores persisted after detachment of the SWI/SNF complex by competition (lane 4). Thus, transient action of the SWI/SNF complex can create stably altered nucleosome cores, suggesting that the complex may act catalytically, an important observation in light of the low abundance of the complex in vivo (24).
SWI/SNF-Treated Nucleosome Cores Remain Bound by GAL4-AH After SWI/SNF Detachment.
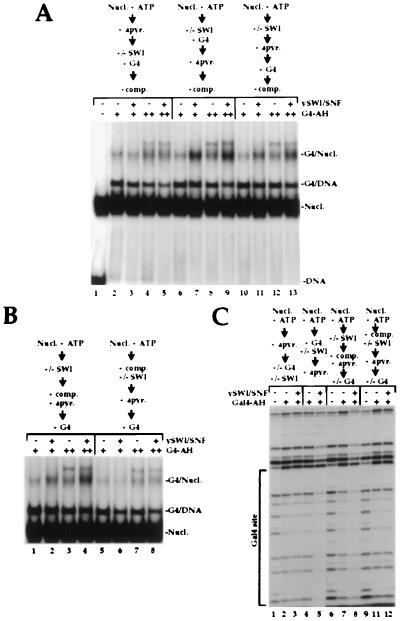
It is well-established that the disruption of nucleosome cores by SWI/SNF is ATP-dependent. Furthermore, ATP hydrolysis is required for SWI/SNF to stimulate transcription factor binding to nucleosome cores (9, 10, 24, 25). More recently, it has been shown that the removal of ATP using apyrase subsequent to SWI/SNF stimulation of factor binding did not reverse the enhanced binding of the factor (29). Thus, ATP is not continuously required for the SWI complex to maintain the stimulated binding of transcription factor to nucleosome core DNA. Because subsequent ATP removal does not detach SWI from nucleosome cores (ATP-independent binding, see above), we determined whether SWI/SNF binding to nucleosome cores is required to preserve the enhanced binding of GAL4-AH to nucleosomal DNA. To test this possibility, we analyzed SWI/SNF stimulation of GAL4-AH binding to a nucleosome core with a single GAL4 site under different regimens of apyrase treatment, GAL4-AH addition and SWI/SNF detachment by competition (Fig. 3A). Apyrase treatment before the addition of SWI/SNF precluded the SWI/SNF-enhanced binding of GAL4-AH to the nucleosome cores (Fig. 3A, compare lanes 2 and 3, 4 and 5). By contrast, if apyrase treatment of the reaction occurred subsequent to the addition of SWI/SNF and GAL4-AH, the stimulation of GAL4-AH binding occurred and survived detachment of the SWI/SNF complex by competitors (compare lanes 6 and 7, 8 and 9). Interestingly, if apyrase treatment followed SWI addition but preceded GAL4 addition, SWI/SNF-dependent enhancement of GAL4-AH binding also was observed (lanes 10–13). These data illustrate that the ATP-dependent disruption of nucleosome cores by the SWI/SNF complex resulted in an altered nucleosome core conformation to which GAL4-AH subsequently could bind in the absence of ATP. More importantly, the SWI/SNF-treated nucleosome cores remain bound by GAL4-AH after SWI/SNF detachment. Thus, stable nucleosome core binding by GAL4-AH not only persisted after the subsequent removal of ATP (as shown previously, ref. 29) but also persisted after the detachment of the SWI/SNF complex.
Figure 3.
Temporal analysis of SWI/SNF action and enhanced activator binding. (A) SWI/SNF-treated nucleosome cores remain bound by GAL4-AH after SWI/SNF detachment. A 154-bp DNA probe bearing a single GAL4 site 32 bp from one end was reconstituted into nucleosome cores and incubated with 30 nM (lanes 2, 3, 6, 7, 10, and 11) and 100 nM (lanes 4, 5, 8, 9, 12, and 13) GAL4-AH dimers. All lanes contain 12.5 nM total nucleosomes and 1 mM ATP, and 10 nM SWI/SNF was included in the odd-numbered lanes. To study the temporal ATP requirement for SWI action, ATP was degraded by apyrase before SWI/SNF addition (lanes 2–5), after SWI/SNF and GAL4 incubation (lanes 6–9), or after SWI/SNF action but before GAL4 incubation (lanes 10–13). Finally, before loading the native gel, nucleosome cores were released from SWI by competition with 20-fold excess of cold oligonucleosomes for 30 min at 30°C. (B) SWI/SNF treatment of nucleosome cores allows GAL4-binding after SWI/SNF detachment. Gel shift assay of similar binding reactions as in A except for the time of SWI/SNF competition. In lanes 1–4 competitors were added after SWI/SNF action, at the same time as apyrase treatment, before GAL4 addition. In lanes 5–8 competitors were included from the beginning of the binding reaction, at the same time as SWI/SNF is added. (C) Footprinting assay of equivalent binding reactions. 30 nM GAL4-AH dimers was added in lanes 2–5 whereas 600 nM was used in lanes 7, 8, 11, and 12. SWI/SNF (40 nM) was added at the indicated time in lanes 3, 5, 8, and 12. Competitors were added at the indicated time in lanes 6–12. Two-fold more DNase I was used in lanes 6–12.
SWI/SNF Treatment of Nucleosome Cores Allows GAL4-AH Binding After SWI/SNF Detachment.
The data in Fig. 3A illustrate that stimulation of transcription factor binding by SWI/SNF could be temporally separated from the ATP-dependent action of the complex. However, it was possible that enhanced nucleosome core binding of GAL4-AH after ATP depletion still required the association of the SWI/SNF complex with the nucleosome core. To test this possibility, we detached SWI/SNF from disrupted nucleosome cores before the addition of GAL4-AH. Detachment of SWI/SNF by a 20-fold molar excess of competitor nucleosomes and simultaneous ATP removal by apyrase before the addition of GAL4-AH still allowed enhanced binding of the activator to nucleosomal DNA (Fig. 3B, compare lanes 1 and 2, 3 and 4). Thus, persistent nucleosome core disruption by the SWI/SNF complex stimulated GAL4-AH binding subsequent to the dissociation of SWI/SNF from the nucleosome core. By contrast, if competitors were added at the same time as SWI/SNF so that efficient SWI/SNF interaction with the GAL4-site nucleosome cores was prevented, no subsequent stimulation of GAL4 binding was detected (Fig. 3B, lanes 5–8). This observation indicates that enhanced activator binding required the interaction of SWI/SNF with the target nucleosome cores but did not require the simultaneous binding of SWI/SNF and GAL4-AH to the nucleosome cores. This result is further illustrated in the DNase footprinting reactions shown in Fig. 3C. Again enhanced GAL4-AH binding was detected subsequent to SWI/SNF detachment from the GAL4-site nucleosome cores (lanes 7 and 8) as long as SWI/SNF was permitted to interact with the target nucleosome cores in the presence of ATP before the addition of competitors (see control lanes 2 and 3, 10 and 11).
The data presented in Figs. 2 and 3 illustrate that the disrupted state of the nucleosome core survived SWI/SNF detachment (Fig. 2) and that nucleosome cores treated with SWI/SNF in the presence of ATP retain an enhanced affinity for GAL4-AH after SWI/SNF detachment (Fig. 3 B and C). Together these data indicate that it is the disrupted state of the nucleosome core that is responsible for the increased affinity of the activator rather than any direct interaction between the activator and the SWI/SNF complex. Thus, these experiments illustrate a temporal separation between SWI/SNF action and enhanced activator binding, suggesting that transient action of the SWI/SNF complex in chromatin can enhance subsequent binding of transcription factors. This interpretation is in agreement with the fact that SWI/SNF is able to stimulate the binding of different transcription factors to nucleosomal DNA (i.e., GAL4, NF-κB, Sp1, and USF; ref. 26).
The SWI/SNF-Disrupted State of Nucleosome Cores Can Persist for Several Hours Before Reverting Back to the Initial Structure.
Because our earlier studies detected reversible changes in apparent nucleosome-positioning mediated by the SWI/SNF complex (27, 28), we tested the persistence of the SWI/SNF-disruption of rotational phasing of nucleosomal DNA after SWI/SNF detachment (Fig. 4). After an initial incubation in presence of SWI/SNF and ATP, both were removed by adding apyrase and excess nonspecific nucleosomes and DNA (20-fold excess of each); incubation was shifted to 37°C and DNase I was added at different time points. In the absence of SWI/SNF, the nucleosome core structure remained unchanged under those conditions over a period of 24 hr (Fig. 4, lanes 1–8). When SWI/SNF was present, disruption of the 10-bp periodicity of DNase I cutting was again seen (lane 9 compared with 1–8). A time course of DNase I addition during the incubation under competition conditions clearly shows that the disrupted digestion pattern persisted for 4 hr (lanes 9–14 compared with lanes 1–6). However, the initial nucleosomal structure was almost completely recovered after 12 and 24 hr of incubation (compare lanes 15 and 16 to 7 and 8). Because the supply of ATP is degraded within 20 min by apyrase and SWI/SNF is detached from nucleosome cores within 30 min (Figs. 2 and 3), we can conclude that the SWI/SNF disrupted state of nucleosome cores is stable for at least 4 hr after its detachment.
The slow reversibility of the SWI/SNF disruption of nucleosome cores as revealed by a loss of rotational phasing of the DNA differs from the more rapid reversible effects of SWI/SNF described in our earlier reports. The previous studies detected a loss of detectable nucleosome positioning and an increase in accessibility of a SalI site at SWI/SNF action on nucleosome arrays. However, these effects were quickly reversed at detachment of the SWI/SNF complex or depletion of ATP (27, 28). Thus, the observed effects of SWI/SNF on the nucleosome arrays appears to require continued binding and function of the SWI/SNF complex. By contrast, in the higher resolution mononucleosomes studies presented here the disruption of rotational phasing of DNA on the surface of the histone octamer can persist for several hours after detachment of the SWI/SNF complex and ATP depletion (Fig. 4). There are at least two possible reasons for this apparent difference. (i) Nucleosome arrays may mediate a more rapid reversal of the SWI/SNF disrupted conformation (e.g., via nucleosome/nucleosome interactions). (ii) The low-resolution nuclease digestion assays used on nucleosome arrays may reveal different features of SWI/SNF disruption than that revealed by high-resolution assays on mononucleosomes. However, the level of nucleosome core disruption revealed by the loss of rotational phasing is sufficient to maintain the increased accessibility of nucleosomal DNA to transcription factors (Fig. 3 B and C) and, thus, is likely to be important in either context and presumably in vivo. We therefore, sought to examine further the nature of the SWI/SNF nucleosome core disruption, which is revealed by the loss of rotational phasing.
SWI/SNF Alters the Path of DNA Bending Around the Histone Octamer.
To further analyze the stably altered nucleosome core conformation generated by the SWI/SNF complex, we investigated further the change in nucleosomal DNase I cutting patterns resulting from SWI/SNF action (Figs. 2B and 4). The stimulation of GAL4-AH binding by the SWI/SNF complex resulted in the release of a ternary complex containing histones, DNA and GAL4-AH (GAL4/Nucl; Fig. 3 A and B). Thus, the action of SWI/SNF in enhancing GAL4-AH binding did not apparently dislodge the histones from the DNA fragment. This observation suggests that although the action of SWI/SNF altered the DNase I digestion pattern of the rotationally positioned nucleosome cores to more closely resemble that of naked DNA (24), the DNA actually remained associated with the core histones.
The repeating pattern of DNase I cutting and protection observed on the end-labeled nucleosome cores in Fig. 2B resulted from the fact that the 5S DNA adopts a preferred path of DNA bending around the histone octamer. This bending generates a sequence-specific “rotational phasing” of the DNA helix where a specific side faces the solvent, and is cut in the minor groove by DNase I, whereas the other side faces the histone octamer, and the minor groove is protected. SWI/SNF could disrupt this pattern of DNase I cleavage without displacing the histone octamer if its action perturbed the preferred orientation of DNA bending, thereby scrambling the rotational phasing of the DNA helix. In this instance, a more random pattern of cutting would occur because the reconstituted nucleosome cores would present a heterogeneous mixture of DNA helix orientations (rotational frames) on the surface of the histone octamer. However, if the DNA helix remained associated with the histone octamer, each individual DNA molecule should retain a 10- to 11-bp repeating pattern of DNase I sensitivity, albeit in a sequence-independent manner. To test this possibility, we reconstituted nucleosome cores with 5S DNA fragments that were labeled throughout the body of the fragment by PCR (body labeled). Digestion of these nucleosome cores with DNase I to an average of two or more cuts per fragment should reveal the distance between two DNase I cuts (as opposed to the distance to the original end of the fragment) independent of the orientation of the DNA sequence on the octamer surface.
Analysis of the effect of the SWI/SNF complex on the DNase I digestion pattern of body-labeled 5S nucleosome cores is shown in Fig. 5. Under DNase I digestion conditions where the majority of the digestion products are larger than 80 bp, a nucleosomal 10-bp ladder is not readily apparent (Fig. 5A, lanes 6–9). This is in contrast to the end-labeled probe where a repeating pattern is easily seen (lanes 3 and 4). However, as digestion of the body-labeled nucleosome cores proceeded to the point where most digestion products represented the distance between two separate DNase I cuts on the nucleosome cores, a 10-bp ladder appeared (lane 10). The body-labeled 5S nucleosome cores then were treated in the presence or the absence of the SWI/SNF complex and ATP (Fig. 5B, lanes 2–5). In contrast to the end-labeled probe (lanes 7–10), the action of SWI/SNF on the body-labeled nucleosome cores in presence of ATP did not result in a disruption of the DNase I digestion pattern (although SWI/SNF did modestly effect the level of digestion). Indeed, even three times more SWI/SNF complex than that used for effective disruption of the end-labeled nucleosome cores did not disrupt the DNase I digestion pattern of the body-labeled 5S nucleosome cores (Fig. 5C; lane 4). These data show that SWI/SNF perturbs the path of DNA bending around the histone octamer.
DISCUSSION
The present study advances our understanding of the functions of the SWI/SNF complex in several aspects. (i) These data demonstrate direct high affinity binding of the SWI/SNF complex to nucleosome cores and that this binding is required for ATP-dependent nucleosome core disruption. (ii) ATP-dependent nucleosome core disruption by the SWI/SNF complex includes a perturbation of the preferred path of DNA bending around the histone octamer (i.e., rotational phasing) that does not require a loss of the histone proteins. (iii) The SWI/SNF-disrupted nucleosome core conformation persists after the detachment of the SWI/SNF complex but eventually can revert back to the original nucleosome core conformation. (iv) The disrupted nucleosome core conformation retains an enhanced affinity for transcription factors after detachment of the SWI/SNF complex, temporally separating nucleosome core disruption by SWI/SNF and stimulated activator binding.
Rotational phasing of DNA sequences on nucleosome cores is thought to largely result from intrinsic curvatures or anisotrophic flexibility of DNA sequences, which enhances the affinity of DNA sequences for histone octamers (32). Thus, the action of SWI/SNF in stimulating factor binding might be explained by the complex reducing the curvature of DNA sequences around the nucleosome core, thereby, reducing the strength of DNA interactions with the histone octamer and enhancing the affinity of DNA sites for transcription factors. Such a model is consistent with the observation that the human SWI/SNF complex reduces the number of stable supercoils in nucleosome-assembled plasmid DNA (10, 29) and is likely related to the DNA binding properties of SWI/SNF, which resembles that of high mobility group-box proteins and can induce positive supercoils in naked DNA (in the presence of Escherichia coli topoisomerase I) (31). A similar interaction of SWI/SNF with nucleosomal DNA might require the energy of ATP hydrolysis to effect an allosteric change in nucleosome core structure. Importantly, this does not seem to involve a helicase activity of the SWI/SNF complex. The purified SWI/SNF complex does not appear to contain helicase activity (24) and single-stranded regions are not detectable in SWI/SNF disrupted nucleosome cores (J.C. and J.L.W., unpublished data).
The continued accumulation of mechanistic data prompts us to revise our previously proposed model for SWI/SNF disruption of nucleosome cores and stimulation of activator binding. Earlier we suggested that SWI/SNF action might provoke the loss of one or two H2A/H2B dimers from the core particle, stabilizing Gal4-AH-nucleosome core interaction (2, 24). However, the data presented here showing the reversibility of the disrupted nucleosome core conformation argues against a loss of histone components during SWI/SNF disruption. These data suggest that SWI/SNF disrupts nucleosome cores primarily by perturbing histone/DNA interactions altering the path of the DNA around the core particle without eviction of histones. In addition to SWI/SNF disruption, actual displacement of histones appears to require further nucleosome core destabilization as provided by the binding of multiple GAL4-AH dimers (27).
The size and subunit complexity of the SWI/SNF complex suggest that it may perform multiple functions and that its in vivo activity may differ in important details from the activities observed in vitro thus far. For example, the mechanisms by which the SWI/SNF complex might be targeted to specific chromosomal loci remains a topic of intense interest. Likely candidates for involvement in targeting of the complex includes interactions that have been detected between the SWI/SNF complex and holo-RNA polymerase (33), the glucocorticoid receptor (34), the retinoblastoma protein (35), and HIV-1 integrase (3). The biochemical studies presented here and in earlier reports support the notion that once recruited to a promoter or enhancer, the SWI/SNF complex has the potential to enhance nucleosome binding by a wide range of transcription factors (26). Enhanced nucleosome binding by transcription factors can occur subsequent to SWI/SNF binding and nucleosome disruption because of the persistence of the disrupted state of the nucleosome core. This observation clearly illustrates that the stimulation of transcription factor binding is caused by the disrupted state of the nucleosome core and does not require transcription factor-SWI/SNF interactions. Thus, the ATP requirement for SWI/SNF function in vitro is to mediate disruption of the nucleosome cores and is only indirectly required to stimulate transcription factor binding. The persistence of the SWI/SNF-disrupted nucleosome core conformation can provide a window of opportunity for enhanced transcription factor binding that extends beyond the actual interaction of the SWI/SNF complex. In the in vitro system used here the disrupted state of the nucleosome cores persisted for up to 4 hr before reverting to the original conformation with a low affinity for transcription factors. The persistence of the SWI/SNF-disrupted nucleosome conformation may be a regulated event in vivo. For example, the reversibility of the disrupted conformation might be enhanced by transcriptional repressors or nucleosome-nucleosome interaction. Moreover, histone modifications though to be involved in transcriptional activity (i.e., histone acetylation) might extend the persistence of the SWI/SNF-disrupted nucleosome conformation. These issues are currently under investigation.
Acknowledgments
We are grateful to Tom Owen-Hughes for pBend5S(172). This work was supported by grants from the National Institute of General Medical Sciences (to J.L.W.) and Canadian Medical Research Council (to J.C.). J.C. was supported by Canadian Medical Research Council fellowships. J.L.W. was a Leukemia Scholar and is currently an Howard Hughes Medical Institute Investigator. C.L.P. is a Leukemia Society Scholar.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Carlson M, Laurent B C. Curr Opin Cell Biol. 1994;6:396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 2.Peterson C L, Tamkun J W. Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 3.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 4.Eisen J A, Sweder K S, Hanawalt P C. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang W, Xue Y, Zhou S, Kuo A, Cairns B, Crabtree G R. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 6.Muchardt C, Yaniv M. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiba H, Muramatsu M, Nomoto A, Kato H. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. Nature (London) 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Côté J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, et al. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nature (London) 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 11.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 12.Dingwall A K, Beek S J, McCallum C M, Tamkun J W, Kalpana G V, Goff S P, Scott M P. Mol Biol Cell. 1995;6:777–791. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elfring L K, Deuring R, McCallum C M, Peterson C L, Tamkun J W. Mol Cell Biol. 1994;14:2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laurent B C, Yang X, Carlson M. Mol Cell Biol. 1992;12:1893–1902. doi: 10.1128/mcb.12.4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsukiyama T, Daniel C, Tamkun J, Wu C. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 16.Tsukiyama T, Wu C. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 17.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 18.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 19.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Nature (London) 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 20.Kruger W, Herskowitz I. Mol Cell Biol. 1991;11:4135–4146. doi: 10.1128/mcb.11.8.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruger W, Peterson C L, Sil A, Coburn G, Arents G, Moudrianakis E N, Herskowitz I. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 22.Hirschhorn J N, Brown S A, Clark C D, Winston F. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 23.Hirschhorn J N, Bortvin A L, Ricupero-Hovasse S L, Winston F. Mol Cell Biol. 1995;15:1999–2009. doi: 10.1128/mcb.15.4.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Côté J, Quinn J, Workman J L, Peterson C L. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 25.Imbalzano A N, Kwon H, Green M R, Kingston R E. Nature (London) 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 26.Utley R T, Côté J, Owen-Hughes T, Workman J L. J Biol Chem. 1997;272:12642–12649. doi: 10.1074/jbc.272.19.12642. [DOI] [PubMed] [Google Scholar]
- 27.Owen-Hughes T, Utley R T, Côté J, Peterson C L, Workman J L. Science. 1996;273:513–516. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- 28.Logie C, Peterson C L. EMBO J. 1997;16:6772–6782. doi: 10.1093/emboj/16.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imbalzano A N, Schnitzler G R, Kingston R E. J Biol Chem. 1996;271:20726–20733. doi: 10.1074/jbc.271.34.20726. [DOI] [PubMed] [Google Scholar]
- 30.Côté J, Utley R T, Workman J L. Methods Mol Genet. 1995;6:108–128. [Google Scholar]
- 31.Quinn J, Fyrberg A M, Ganster R W, Schmidt M C, Peterson C L. Nature (London) 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 32.Crothers D M, Gartenberg M R, Shrader T E. Methods Enzymol. 1991;208:118–147. doi: 10.1016/0076-6879(91)08011-6. [DOI] [PubMed] [Google Scholar]
- 33.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 34.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Science. 1992;258:1598–1603. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 35.Dunaief J L, Strober B E, Guha S, Khavari P A, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]