Abstract
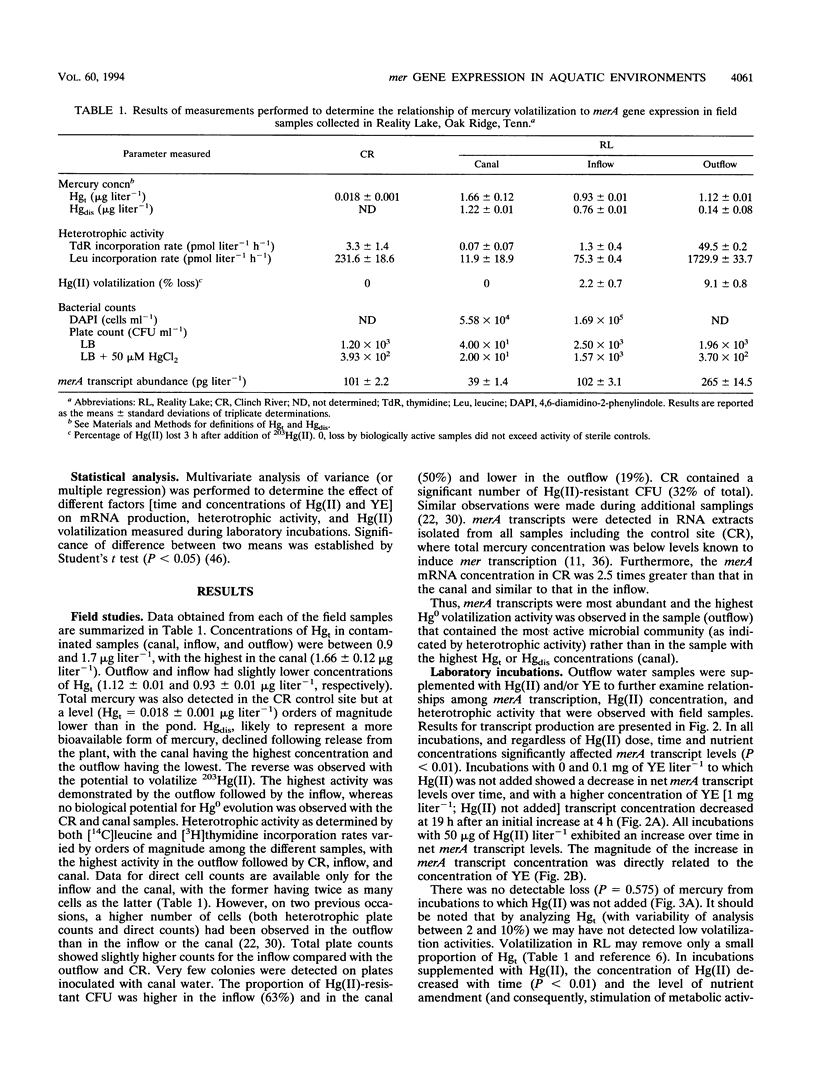
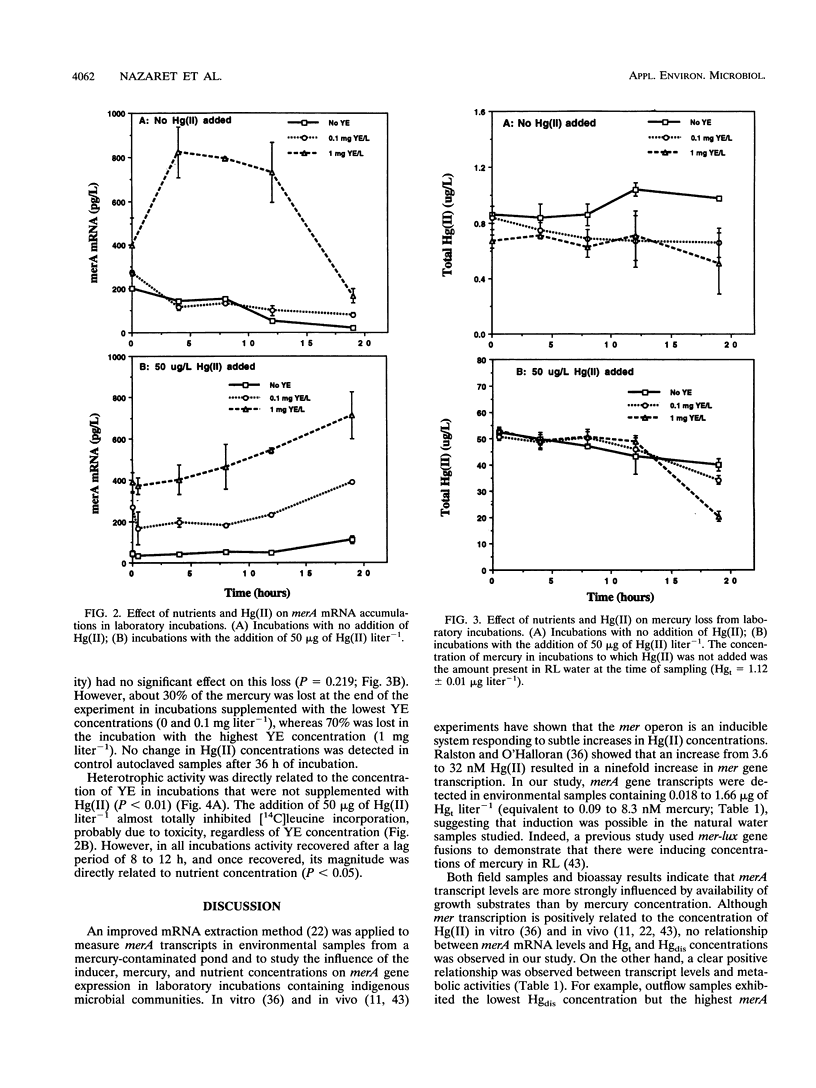
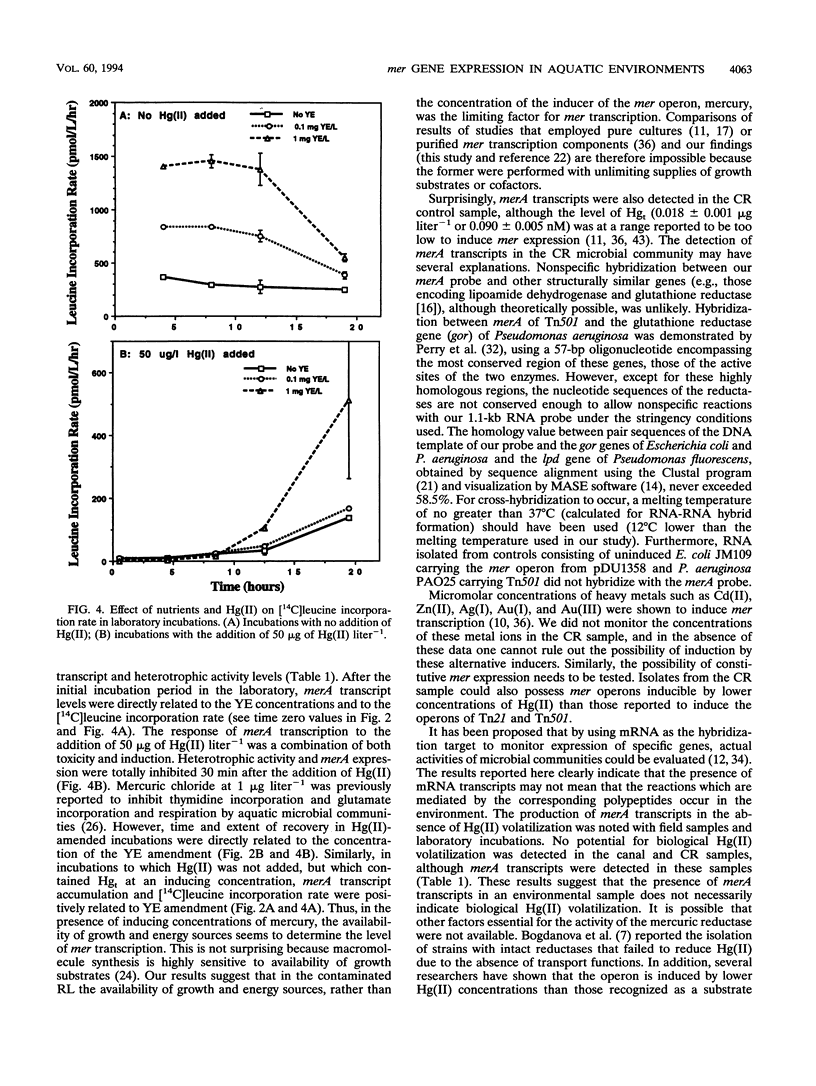
The relationship of merA gene expression (specifying the enzyme mercuric reductase) to mercury volatilization in aquatic microbial communities was investigated with samples collected at a mercury-contaminated freshwater pond, Reality Lake, in Oak Ridge, Tenn. Levels of merA mRNA transcripts and the rate of inorganic mercury [Hg(II)] volatilization were related to the concentration of mercury in the water and to heterotrophic activity in field samples and laboratory incubations of pond water in which microbial heterotrophic activity and Hg(II) concentration were manipulated. Levels of merA-specific mRNA and Hg(II) volatilization were influenced more by microbial metabolic activity than by the concentration of mercury. merA-specific transcripts were detected in some samples which did not reduce Hg(II), suggesting that rates of mercury volatilization in environmental samples may not always be proportional to merA expression.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barkay T., Gillman M., Liebert C. Genes encoding mercuric reductases from selected gram-negative aquatic bacteria have a low degree of homology with merA of transposon Tn501. Appl Environ Microbiol. 1990 Jun;56(6):1695–1701. doi: 10.1128/aem.56.6.1695-1701.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T., Liebert C., Gillman M. Environmental significance of the potential for mer(Tn21)-mediated reduction of Hg2+ to Hg0 in natural waters. Appl Environ Microbiol. 1989 May;55(5):1196–1202. doi: 10.1128/aem.55.5.1196-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T., Liebert C., Gillman M. Hybridization of DNA probes with whole-community genome for detection of genes that encode microbial responses to pollutants: mer genes and Hg2+ resistance. Appl Environ Microbiol. 1989 Jun;55(6):1574–1577. doi: 10.1128/aem.55.6.1574-1577.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanova E. S., Mindlin S. Z., Pakrová E., Kocur M., Rouch D. A. Mercuric reductase in environmental gram-positive bacteria sensitive to mercury. FEMS Microbiol Lett. 1992 Oct 1;76(1-2):95–100. doi: 10.1016/0378-1097(92)90370-4. [DOI] [PubMed] [Google Scholar]
- Chin-Leo G., Kirchman D. L. Estimating bacterial production in marine waters from the simultaneous incorporation of thymidine and leucine. Appl Environ Microbiol. 1988 Aug;54(8):1934–1939. doi: 10.1128/aem.54.8.1934-1939.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L., Mukhopadhyay D., Yu H., Kim K. S., Misra T. K. Regulation of the Staphylococcus aureus plasmid pI258 mercury resistance operon. J Bacteriol. 1992 Nov;174(21):7044–7047. doi: 10.1128/jb.174.21.7044-7047.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condee C. W., Summers A. O. A mer-lux transcriptional fusion for real-time examination of in vivo gene expression kinetics and promoter response to altered superhelicity. J Bacteriol. 1992 Dec;174(24):8094–8101. doi: 10.1128/jb.174.24.8094-8101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb R. W., Wagner-Döbler I. Detection of polychlorinated biphenyl degradation genes in polluted sediments by direct DNA extraction and polymerase chain reaction. Appl Environ Microbiol. 1993 Dec;59(12):4065–4073. doi: 10.1128/aem.59.12.4065-4073.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner D. V., Jurka J. Multiple aligned sequence editor (MASE). Trends Biochem Sci. 1988 Aug;13(8):321–322. doi: 10.1016/0968-0004(88)90129-6. [DOI] [PubMed] [Google Scholar]
- Fox B. S., Walsh C. T. Mercuric reductase: homology to glutathione reductase and lipoamide dehydrogenase. Iodoacetamide alkylation and sequence of the active site peptide. Biochemistry. 1983 Aug 16;22(17):4082–4088. doi: 10.1021/bi00286a014. [DOI] [PubMed] [Google Scholar]
- Fox B., Walsh C. T. Mercuric reductase. Purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J Biol Chem. 1982 Mar 10;257(5):2498–2503. [PubMed] [Google Scholar]
- Gambill B. D., Summers A. O. Synthesis and degradation of the mRNA of the Tn21 mer operon. J Mol Biol. 1992 May 20;225(2):251–259. doi: 10.1016/0022-2836(92)90919-b. [DOI] [PubMed] [Google Scholar]
- Gilbert M. P., Summers A. O. The distribution and divergence of DNA sequences related to the Tn21 and Tn501 mer operons. Plasmid. 1988 Sep;20(2):127–136. doi: 10.1016/0147-619x(88)90015-7. [DOI] [PubMed] [Google Scholar]
- Hahn D., Amann R. I., Zeyer J. Whole-Cell Hybridization of Frankia Strains with Fluorescence- or Digoxigenin-Labeled, 16S rRNA-Targeted Oligonucleotide Probes. Appl Environ Microbiol. 1993 Jun;59(6):1709–1716. doi: 10.1128/aem.59.6.1709-1716.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Jeffrey W. H., Nazaret S., Von Haven R. Improved Method for Recovery of mRNA from Aquatic Samples and Its Application to Detection of mer Expression. Appl Environ Microbiol. 1994 Jun;60(6):1814–1821. doi: 10.1128/aem.60.6.1814-1821.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K. F., Pedersen S. Metabolic growth rate control in Escherichia coli may be a consequence of subsaturation of the macromolecular biosynthetic apparatus with substrates and catalytic components. Microbiol Rev. 1990 Jun;54(2):89–100. doi: 10.1128/mr.54.2.89-100.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas R. B., Gilmour C. C., Stoner D. L., Weir M. M., Tuttle J. H. Comparison of methods to measure acute metal and organometal toxicity to natural aquatic microbial communities. Appl Environ Microbiol. 1984 May;47(5):1005–1011. doi: 10.1128/aem.47.5.1005-1011.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P. A., Ford S. J., Brown N. L. Transcriptional regulation of the mercury-resistance genes of transposon Tn501. J Gen Microbiol. 1986 Feb;132(2):465–480. doi: 10.1099/00221287-132-2-465. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Sakamoto M., Uchiyama H., Yagi O. Organomercurial-volatilizing bacteria in the mercury-polluted sediment of Minamata Bay, Japan. Appl Environ Microbiol. 1990 Jan;56(1):304–305. doi: 10.1128/aem.56.1.304-305.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry A. C., Ni Bhriain N., Brown N. L., Rouch D. A. Molecular characterization of the gor gene encoding glutathione reductase from Pseudomonas aeruginosa: determinants of substrate specificity among pyridine nucleotide-disulphide oxidoreductases. Mol Microbiol. 1991 Jan;5(1):163–171. [PubMed] [Google Scholar]
- Pichard S. L., Paul J. H. Gene expression per gene dose, a specific measure of gene expression in aquatic microorganisms. Appl Environ Microbiol. 1993 Feb;59(2):451–457. doi: 10.1128/aem.59.2.451-457.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichard Scott L., Paul John H. Detection of Gene Expression in Genetically Engineered Microorganisms and Natural Phytoplankton Populations in the Marine Environment by mRNA Analysis. Appl Environ Microbiol. 1991 Jun;57(6):1721–1727. doi: 10.1128/aem.57.6.1721-1727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt D., Abshagen U., Mühlberg W., Horn H. J., Schmitt-Rüth R., Vollmar J. The influence of age and multimorbidity on the pharmacokinetics and metabolism of spironolactone. Arch Gerontol Geriatr. 1984 Jul;3(2):147–159. doi: 10.1016/0167-4943(84)90006-2. [DOI] [PubMed] [Google Scholar]
- Ralston D. M., O'Halloran T. V. Ultrasensitivity and heavy-metal selectivity of the allosterically modulated MerR transcription complex. Proc Natl Acad Sci U S A. 1990 May;87(10):3846–3850. doi: 10.1073/pnas.87.10.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinderle S. J., Booth J. E., Williams J. W. Mercuric reductase from R-plasmid NR1: characterization and mechanistic study. Biochemistry. 1983 Feb 15;22(4):869–876. doi: 10.1021/bi00273a025. [DOI] [PubMed] [Google Scholar]
- Rochelle Paul A., Wetherbee Mary K., Olson Betty H. Distribution of DNA Sequences Encoding Narrow- and Broad-Spectrum Mercury Resistance. Appl Environ Microbiol. 1991 Jun;57(6):1581–1589. doi: 10.1128/aem.57.6.1581-1589.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selifonova O., Burlage R., Barkay T. Bioluminescent sensors for detection of bioavailable Hg(II) in the environment. Appl Environ Microbiol. 1993 Sep;59(9):3083–3090. doi: 10.1128/aem.59.9.3083-3090.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O. Organization, expression, and evolution of genes for mercury resistance. Annu Rev Microbiol. 1986;40:607–634. doi: 10.1146/annurev.mi.40.100186.003135. [DOI] [PubMed] [Google Scholar]
- Summers A. O. Untwist and shout: a heavy metal-responsive transcriptional regulator. J Bacteriol. 1992 May;174(10):3097–3101. doi: 10.1128/jb.174.10.3097-3101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Effects of Hg, CH(3)-Hg, and Temperature on the Expression of Mercury Resistance Genes in Environmental Bacteria. Appl Environ Microbiol. 1990 Nov;56(11):3266–3272. doi: 10.1128/aem.56.11.3266-3272.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Park M. J., Olson B. H. Rapid method for direct extraction of mRNA from seeded soils. Appl Environ Microbiol. 1991 Mar;57(3):765–768. doi: 10.1128/aem.57.3.765-768.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]



