Abstract
Switching of the transcription pattern in Escherichia coli during the growth transition from exponential to stationary phase is accompanied by the replacement of the RNA polymerase-associated σ70 subunit (σD) with σ38 (σS). A fraction of the σ70 subunit in stationary phase cell extracts was found to exist as a complex with a novel protein, designated Rsd (Regulator of sigma D). The intracellular level of Rsd starts to increase during the transition from growing to stationary phase. The rsd gene was identified at 90 min on the E. coli chromosome. Overexpressed and purified Rsd protein formed complexes in vitro with σ70 but not with other σ subunits, σN, σS, σH, σF, and σE. Analysis of proteolytic fragments of σ70 indicated that Rsd binds at or downstream of region 4, the promoter −35 recognition domain. The isolated Rsd inhibited transcription in vitro to various extents depending on the promoters used. We propose that Rsd is a stationary phase E. coli protein with regulatory activity of the σ70 function.
Keywords: transcriptase control, stationary phase control, enzyme activity control, RNA polymerase storage
The RNA polymerase of Escherichia coli is composed of core enzyme with the subunit structure α2ββ′ combined with one of seven different species of σ subunit. The core enzyme carries all the catalytic functions for RNA polymerization, but a σ subunit is required for the proper initiation of transcription at promoters (1–3). The number of core enzyme molecules in an E. coli cell is approximately 2,000 (4, 5), whereas there are more than 4,000 genes on the E. coli chromosome to be transcribed at widely varying rates (5, 6). The σ subunit required for transcription of most of the genes expressed in the exponential growth phase is σ70, the product of the rpoD gene (1, 2). Its concentration during exponential phase is about 700 molecules per cell, as measured by immunoprecipitation of radiolabeled cell extracts (7) or Western blot analysis with specific antibodies (8, 9). In growing cells, at any given moment, about two-thirds (about 1,300–1,400 molecules) of the core enzyme is actively involved in elongation of transcripts. In this state, the core enzyme is bound to DNA but not associated with σ. Accordingly most of the remaining core enzyme (about 600–700 molecules) is likely to be associated with σ70 (5, 10).
Upon entry into stationary phase, σ38 or σS (the product of the rpoS gene) begins to be produced and allows core polymerase to recognize and transcribe the genes required for stationary phase survival (11–14). The level of σS increases to as much as about 250–300 molecules per cell (about 30–35% the level of σ70) (8, 9). Meanwhile the level of σ70 stays constant, even though the frequency of transcription of genes under the control of σ70 decreases by more than 10-fold (5). The synthesis of σS is also induced on exposure of growing cells to certain stresses such as high osmolarity or high temperature, and it plays a role in transcription of some stress-response genes (14, 15). Known factors that contribute to the preferential utilization of σS rather than σ70 in the stationary phase or under stress conditions include increases in intracellular concentrations of potassium glutamate, trehalose, glycine betaine, and polyphosphate and a decrease in DNA superhelicity, all of which tend to reduce the activity of Eσ70 but increase that of EσS (16).
In addition, we found in this study that in stationary phase cells, σ70 but not other σ subunits forms a complex with a hitherto unidentified stationary-specific protein. This protein begins to be synthesized during the transition from exponential growth to stationary phase. In vitro transcription studies indicated that the protein interferes with the engagement of σ70 in the transcription cycle. After sequencing, the protein was identified as a product of the unidentified reading frames revealed by genome sequence analysis (17). We propose that this protein, designated Rsd (Regulator of sigma D), with σ70 binding activity plays a role in controlling the σ70 function in stationary phase E. coli.
MATERIALS AND METHODS
Expression Plasmids for the σ Subunits, FlgM and Rsd.
To create plasmids for expression in E. coli of various σ subunits in glutathione S-transferase (GST) fusion form, the respective coding sequences were PCR-amplified with the primers described in Table 1. Each primer includes a unique restriction enzyme site (underlined in the sequences shown in Table 1). The sequences of the PCR-amplified fragments were confirmed by the dideoxynucleotide method. The PCR-generated DNA fragments were treated with the appropriate restriction enzymes and cloned into the corresponding sites of the pGEX5x-1 vector (Pharmacia) to generate pGEXD, pGEXN, pGEXS, pGEXH, pGEXF, pGEXM, and pGEXRD for expression of GST fusion proteins of σ70, σN (σ54), σS (σ38), σN (σ32), σF (σ28), FlgM (anti-σF), and Rsd, respectively. The rsd gene was also inserted into pET21a (Novagen) between the NdeI and XhoI sites to generate pET-Rsd, for expression of His6-tagged Rsd protein.
Table 1.
Primers used for gene amplification
| Gene | Primer | Sequence |
|---|---|---|
| rpoD (σ70 gene) | D1 | 5′-CCCAAGCTTGAATTCATGGAGCAAAACCCGCAGTCAC-3′ |
| D4 | 5′-CCCAAGCTTGTCGACTTAATCGTCCAGGAAGCTACG-3′ | |
| rpoN (σ54 gene) | N1 | 5′-GGAATTCCATATGAAGCAAGGTTTGCAACTCAGG-3′ |
| N2 | 5′-CATGCATGCCTCGAGTCAAACGAGTTGTTTACGCTG-3′ | |
| rpoS (σ38 gene) | S5 | 5′-GCGAATTCCATATGTTCCGTCAAGGGATCACG-3′ |
| S6 | 5′-GCGGATCCCTCGAGTTACTCGCGGAACAGCGCTTC-3′ | |
| rpoF (σ28 gene) | F1 | 5′-CGAGCTCGGATCCCCATGAATTCACTCTATACCGCT-3′ |
| F2 | 5′-GGGGTACCCTCGAGTTATAACTTACCCAGTTTAGT-3′ | |
| flgM (FlgM gene) | M1 | 5′-GCGAATTCCATATGAGTATTGATCGCACTTCGCC-3′ |
| M2 | 5′-GCGGATCCCTCGAGGTTACTCTGCAAGTCTTGCTG-3′ | |
| rsd (Rsd gene) | RsdU | 5′-GCGAATTCCATATGCTTAACCAGCTCGATAACC-3′ |
| RsdR | 5′-AACTGCAGCTCGAGAGCAGGATGTTTGACGCGGGC-3′ |
Purification and Analysis of σ-Associated Proteins.
To identify σ-associated proteins, an E. coli W3110 type A strain (18) containing pGEXD, pGEXN, pGEXS, or pGEXF was grown in Luria–Bertani (LB) medium containing ampicillin (100 μg/ml) at 37°C until stationary phase (3–4 h after cessation of cell growth). Cell lysates were prepared essentially as described previously (19) and applied onto a glutathione–Sepharose column (Pharmacia) previously equilibrated with PBS. The column was washed with 10× bed volumes of PBS and eluted with glutathione-containing buffer (50 mM glutathione/100 mM Tris⋅HCl, pH 8.0/120 mM NaCl). Aliquots of the pooled eluate fractions were fractionated by electrophoresis on 7.5, 10, and 13.5% polyacrylamide gels in the presence of SDS. Proteins were transferred onto PVDF membranes (Nippon Genetics) and stained with Coomassie brilliant blue. Stained protein bands were directly subjected to automated microsequencing (Applied Biosystems 491 protein sequencer). Purification and analysis of anti-σ-associated proteins were carried out by the same procedure by using pGEXM and pGEXRD.
Preparation of σ Subunits and Rsd.
Overexpression and purification of σ70, σN, σS, σH, σF, and σE were performed as described previously (19–21). To overproduce the His6-tagged or GST-fused Rsd proteins, an E. coli BL21(λDE3) transformant containing pET-Rsd or a strain DH5 transformant containing pGEX-Rsd was grown at 37°C in LB medium containing ampicillin, and when the culture reached 30 Klett units (540-nm filter), isopropyl β-d-thiogalactopyranoside was added at 1 mM. After 1 h, cells were harvested and stored at −80°C until use. Cell lysates were prepared essentially as described previously (19). For the purification of His6-tagged Rsd protein, the soluble fraction was applied onto a Ni2+-nitrilotriacetic acid column (Qiagen) equilibrated with a binding buffer (20 mM Tris⋅HCl, pH 8.0/0.5 M NaCl). The column was washed with 10× bed volumes of the binding buffer containing 5 mM imidazole, followed by elution with a step gradient of imidazole from 10 to 200 mM. The eluate containing the Rsd protein was dialyzed against TGED buffer (10 mM Tris⋅HCl, pH 7.6/5% glycerol/0.1 mM EDTA/0.1 mM DTT/0.1 M NaCl) and loaded onto a heparin–agarose column (Hi-Trap Heparin, Pharmacia) equilibrated with TGED buffer. The adsorbed proteins were eluted with a 0.1–1.0 M linear gradient of NaCl in TGED. The Rsd protein, which eluted at 0.8 M NaCl, was dialyzed against storage buffer (10 mM Tris⋅HCl, pH 7.6/10 mM MgCl2/0.2 M KCl/50% glycerol/0.1 mM EDTA/1 mM DTT). The purification of GST–Rsd protein was carried out essentially as described above.
Antibodies Against σ Subunits and Rsd.
Antibodies against each σ subunit or anti-σ were produced in rabbits by injecting the respective purified proteins. No significant crossreaction was observed for the antibodies used in this study.
GST Pull Down Assay.
For analysis of σ and Rsd interaction, 20 pmol each of GST or GST–Rsd were mixed with equimolar amounts of holoenzyme, core enzyme, or σ subunit on ice for 30 min in a total volume of 50 μl of transcription buffer (50 mM Tris⋅HCl, pH 7.8, at 37°C/3 mM magnesium acetate/50 mM NaCl/0.1 mM EDTA/0.1 mM DTT/25 μg/ml BSA). After adding glutathione–Sepharose beads (Pharmacia) according to the standard procedure recommended by the manufacturer, the mixtures were incubated on ice for 10 min. The beads carrying immobilized proteins were washed 3 times with PBS, and then the bound proteins were eluted with glutathione buffer (50 mM glutathione/100 mM Tris⋅HCl, pH 8.0/120 mM NaCl) and separated by SDS/PAGE on 5–15% gradient gels. The gels were analyzed by Western blotting with polyclonal antibodies against α, β, β′, and each of the σ subunits, as previously described (8, 9).
Limited Digestion with Trypsin.
Limited trypsin digestion of σ70 was carried out in a 125-μl reaction mixture containing 20 mM Tris⋅HCl (pH 8.0), 50 mM NaCl, 5% glycerol, 0.1 mM EDTA, 1 mM DTT, 1 fmol of l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin (Sigma), and 1 nmol of sample protein. The reaction was carried out for 20 min at 25°C and then terminated by adding 5 μl of 100 mM phenylmethylsulfonyl fluoride. Reaction products were analyzed by SDS/PAGE on 5–15% gradient gels.
In Vitro Single-Round Transcription Assay.
RNA polymerase core enzyme was purified from E. coli W3350 by passing the purified RNA polymerase at least three times through phosphocellulose columns (22). Holoenzymes were reconstituted by mixing the core enzyme and 3-fold molar excesses of σ subunits. Single-round transcription by the holoenzyme was carried out under the standard conditions described previously (23) except that the concentrations of templates and RNA polymerase were varied as indicated in each experiment. In brief, mixtures of template and RNA polymerase were preincubated at 37°C for 30 min to allow open complex formation in 35 μl of the standard transcription mixture containing 50 mM NaCl. RNA synthesis was initiated by adding a 15-μl mixture of substrate and heparin in the standard transcription buffer and continued for 5 min at 37°C. Transcripts were precipitated with ethanol and analyzed by electrophoresis on 6 or 8% polyacrylamide gels containing 8 M urea. Gels were exposed to imaging plates, and the plates were analyzed with a BAS-2000 image analyzer (Fuji). The templates used were: a 205-bp EcoRI–EcoRI fragment for lacUV5 (23); a 233-bp HpaII–HpaII fragment for gal (24); a 205-bp PvuII–XbaI fragment for wild-type lac (19); and a 287-bp BamHI-KpnI fragment for the alaS promoter (25). These truncated DNA templates produced in vitro transcripts 63, 45, 68, and 169 nucleotides in length, respectively.
For the anti-σ70 activity assay of Rsd, a fixed amount of σ70 (1 pmol) and increasing amounts of Rsd (1, 2, 5, and 10 pmol) were preincubated for 10 min at 30°C, and then 1 pmol of core enzyme was added. The mixtures were subjected to the single-round transcription assay.
RESULTS
Analysis of σ Subunit-Associated Proteins.
Previously we determined the intracellular concentrations of various σ subunits in E. coli strain MC4100 and W3110 at various phases of cell growth by using a quantitative Western blot assay (8, 9). The level of σ70, the major σ subunit, was found to stay constant at about 600–700 molecules/cell, in both growing and stationary phases. The results were in good agreement with those obtained by a combination of immunoprecipitation and gel electrophoresis of radiolabeled cell extracts of the strains W3350 and B/r (10). These observations raised a question as to why σ70 is largely inactive in stationary phase. In growing cells, most of the σ70 subunit is associated with core enzyme and is involved, through the sigma cycle, in transcription (5). We therefore tried to identify any proteins which might be associated with σ70 or other σ subunits in stationary phase. For this purpose four species of σ subunits, σ70, σN, σS, and σF, were expressed as fusions with GST in E. coli W3110 grown into stationary phase in LB medium at 37°C. The expression levels of GST–σ fusion proteins were analyzed by Western blotting of induced and uninduced cell lysates. Because of the leaky controls, the level of expression of GST–σ70 subunit in the absence of isopropyl β-d-thiogalactopyranoside was as high as that of the chromosome-coded σ70 (data not shown). Thus, to minimize possible artifacts because of overproduction of σ subunits, σ70-associated proteins were analyzed by using uninduced cell extracts.
Cell lysates were passed directly through glutathione–Sepharose columns. The column-bound proteins were eluted with glutathione and separated by SDS/PAGE on 5–15% gradient gels (Fig. 1). All four GST–σ fusion proteins were bound to the columns and eluted with glutathione (the major band in each lane). By immunostaining, the core enzyme subunits, α, β, and β′, were all detected in the column-bound fractions for each of the GST–σ fusion subunits (GST–σ70, GST–σ54, and GST–σ28 lanes) except GST–σS (GST–σ38 lane), indicating that the holoenzymes containing GST-fused σ70, σN, and σF can be retained on the columns, but the GST–σS does not form a stable holoenzyme or the holoenzyme containing GST–σS has a low affinity to the glutathione–Sepharose column.
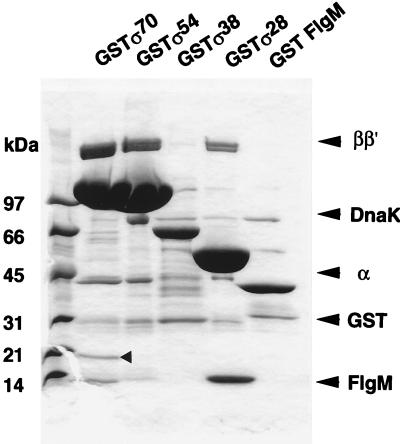
Figure 1.
Identification of σ-associated proteins. Cells of E. coli W3110 containing pGEXD (lane GSTσ70), pGEXN (lane GSTσ54), pGEXS (lane GSTσ38), pGEXF (lane GSTσ28), or pGEXM (lane GST FlgM) were grown in LB medium at 37°C into stationary phase (3–4 h after the cessation of cell growth). Cell lysates were prepared as described under Materials and Methods. GST fusion protein complexes were isolated and separated by SDS/PAGE on a 5–15% gradient gel. The gel was stained with Coomassie brilliant blue. The migration positions of core enzyme subunits, DnaK, GST (without Rsd), and FlgM are indicated on the right. The arrow in the GSTσ70 lane indicates Rsd.
For identification of σ-associated proteins, most of the minor bands detected by protein staining were subjected to protein microsequencing. DnaK was found to be associated with all GST–σ fusion subunits, but its content was the highest for σN. Several minor bands associated with all GST–σ fusions were identified as GST without σ and some other degradation products of GST–σ fusion proteins, because all these proteins contained the N-terminal sequences of GST. In addition, one or two unique proteins were identified for each σ subunit. For instance, the 14 kDa band observed in the bound protein fraction with GST–σF was found to have the protein sequence of FlgM (Fig. 1, GST–σ28 lane), which is the anti-σ factor for σF (26). To verify this finding, we also expressed GST–FlgM fusion and found that σF can be recovered as a complex with the GST–FlgM fusion protein (Fig. 1, GST–FlgM lane). These results confirmed the possibility that as yet unidentified anti-σ factors against other σ subunits could be detected with this experimental system.
Identification of an σ70-Associated Protein (Rsd).
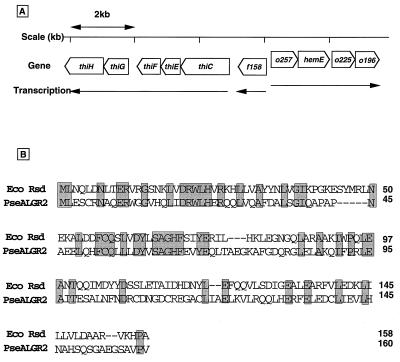
In the GST–σ70 lane, at least five additional bands were observed besides the three core subunits, α, β, and β′ (Fig. 1, lane GST–σ70). After protein microsequencing, four of these were identified as breakdown products of the GST–σ70 fusion protein, and these bands were also observed for other GST–σ fusions. However, one specific band with a molecular mass of around 21 kDa (indicated by an arrow) was identified only in the GST–σ70 lane but not in the other GST lanes. The N-terminal sequence of this 21-kDa protein was found in the E. coli genome database as unidentified reading frame f158 (17). The f158 unidentified reading frame gene is located near 90 min on the E. coli chromosome, probably forming a single gene operon (Fig. 2A).
Figure 2.
Map position of the rsd gene and the amino acid sequence of Rsd protein. (A) Map position of the rsd gene. From the amino acid sequence analysis of Rsd, the rsd gene was found to be identical with the f158 gene in the genome sequence (6). (B) The amino acid sequences of the Rsd protein and the P. aeruginosa AlgR2 protein are compared in optimized alignment. Identical residues are shaded.
We next cloned the f158 gene and confirmed that its nucleotide sequence is identical with that in the E. coli genome database. The predicted gene product shows a 31% identity with the alginate regulatory protein AlgR2 of Pseudomonas aeruginosa (26) (Fig. 2B). More strikingly, there is a 45% identity in its N-terminal region of 74 amino acid residues with the first 69 residues of AlgR2, which regulates the production of alginate, a virulence factor for P. aeruginosa, by controlling some enzymes in the pathway of alginate production such as nucleoside diphosphokinase (27). Because the isolated 21-kDa protein formed binary complexes with σ70 and interfered with its function (see below), we tentatively designated this 21-kDa protein as Rsd.
Direct Interaction of Rsd with σ70 Subunit.
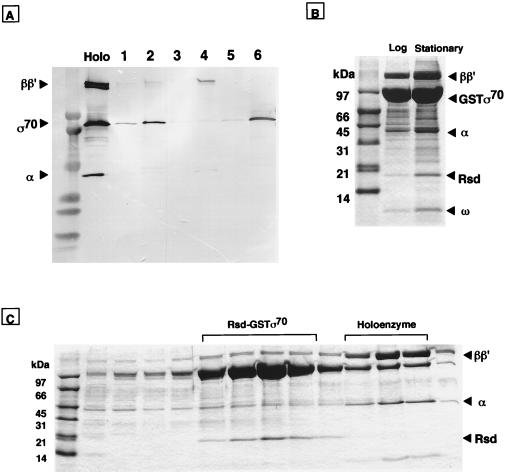
To test whether this σ70-associated Rsd interacts directly with σ70, we constructed an E. coli plasmid expressing a GST–Rsd fusion protein. Overexpressed and purified GST–Rsd was mixed with either σ70 or Eσ70 holoenzyme and then with glutathione–Sepharose beads. Proteins tightly bound to the beads were eluted with glutathione and fractionated by SDS/PAGE. For detection of the proteins at a high sensitivity, the gels were subjected to Western blotting with a mixture of anti-α, anti-β, anti-β′, and anti-σ70 antibodies (Fig. 3A). Although there were low level backgrounds of nonspecific binding to GST without Rsd under the washing conditions employed, it is clear that: free σ70 binds to GST–Rsd (lane 6) but not to GST (lane 5); a fraction of σ70 in the holoenzyme preparation also binds to GST–Rsd (lane 2); but neither α nor ββ′ in both core and holoenzyme associate with GST–Rsd (lanes 4 and 6). These results suggest that Rsd associates preferentially with free σ70.
Figure 3.
Identification of complex formation between σ70 and Rsd. (A) Complex formation in vitro between Rsd and σ70 was analyzed by the GST pull down assay. GST (lanes 1, 3, and 5) or GST–Rsd (lanes 2, 4, and 6) was mixed with equimolar amounts of Eσ70 holoenzyme (lanes 1 and 2), core enzyme (lanes 3 and 4), or σ70 subunit (lanes 5 and 6). Complexes formed were bound to glutathione–Sepharose beads. The bead-bound proteins were eluted with 50 mM glutathione and separated by SDS/PAGE on a 5–15% gradient gel. The gel was subjected to Western blot analysis by using a mixture of monospecific antibodies against RNA polymerase α, β, β′, and σ70 subunits. The migration positions of RNA polymerase subunits can be identified in the holoenzyme lane. (B) Isolation of GST–σ70-associated proteins in cell extracts. Cell lysates of a pGEXD transformant of E. coli W3110 were prepared at both exponential (lane, log) and stationary (lane, stationary) phases. GST–σ70-bound proteins were isolated by the GST pull down assay, and the σ70-bound proteins were separated by SDS/13.5% PAGE. The migration positions of core enzyme subunits, GST–σ70, and ω proteins are indicated on the right. (C) Proteins isolated from stationary phase cells (see stationary lane in B) were subjected to heparin–Sepharose column chromatography. Proteins were eluted by a linear gradient of NaCl, and aliquots were analyzed by SDS/13.5% PAGE.
To identify the intracellular state of Rsd–σ70 complexes, we expressed GST–σ70 in E. coli and analyzed σ70-bound proteins after isolation of complexes with glutathione beads. As shown in Fig. 3B (log lane), the major proteins associated with σ70 in extracts of exponentially growing cells were core enzyme subunits. In addition, several minor bands were detected, which were identified by N-terminal sequencing to be degradation products of GST–σ70. When the stationary phase cell extract was analyzed (Fig. 3B, stationary lane), two additional bands, Rsd and ω, were identified, suggesting that Rsd is produced or becomes active only in the stationary phase cells. The level of RNA polymerase-associated ω protein also increases in stationary phase.
The glutathione bead eluate of stationary phase extracts was then fractionated by heparin–agarose column chromatography. As shown in Fig. 3C, two major peaks were identified, one core-associated GST–σ70 (holoenzyme fractions) and the other free form of GST–σ70, to which Rsd was associated (Rsd–GST–σ70 fractions). The result indicates that Rsd-bound σ70 is unable to associate with core enzyme.
Rsd-Binding Site on σ70 Subunit.
The specificity of σ70 recognition by Rsd was analyzed with six E. coli σ subunits, i.e., σ70, σN, σS, σH, σF, and σE, which were all overexpressed in E. coli and purified to apparent homogeneity. A mixture of six different σ subunits was subjected to the GST–Rsd pull down assay, and the Rsd-bound σ species was identified by immunostaining with monospecific polyclonal antibodies against each of the six σ subunits. Among the six σ subunits examined, only one species, σ70, was found to bind Rsd (Fig. 4A). Because the antibodies against six different σ factors are not equally efficient in binding with their respective σ factors, we repeated the assay with increasing amounts of the GST–Rsd fusion and increasing amounts of the antibodies. Under all the conditions employed, σ70 was the only species tightly bound with Rsd (data not shown). This suggests that Rsd recognizes a unique structural signal of σ70, which is not present in other σ subunits.
Figure 4.
Identification of the Rsd binding subunit and Rsd contact site. (A) GST (lane 1) or GST–Rsd (lane 2) was mixed with an equimolar mixture of σ70, σ54(N), σ38(S), σ32(H), σ28(F), and σ24(E). Complexes were isolated by the GST pull down method with glutathione–Sepharose beads. The bead-bound proteins were eluted with 50 mM glutathione and analyzed by SDS/13.5% PAGE. The gel was subjected to Western blot analysis by using a mixture of antibodies against all six σ subunits. The control lane contained all six σ subunits, which all reacted against the antibody mixture. (B) Trypsin-treated σ70 (starting material, 1 nmol), shown in lane σ70/trypsin, was mixed with two different concentrations of GST–Rsd (lane 1, 40 pmol; lane 2, 20 pmol), and the complexes formed were isolated by the GST pull down assay with glutathione–Sepharose beads. The bead-bound proteins were eluted with 50 mM glutathione and separated by 5–15% SDS/PAGE. The gel was analyzed by Western blotting by using monospecific polyclonal antibodies against σ70. After N-terminal sequence analysis, R3–4 and R4 peptides were identified as C-terminal fragments downstream from 449 and 500, respectively.
We next determined the contact site on σ70 for Rsd. For this purpose, σ70 was partially digested with trypsin, and the digestion products were incubated with either GST or GST–Rsd. Proteins tightly bound to the glutathione–Sepharose beads were eluted with glutathione and analyzed by Western blotting with anti-σ70 antibodies. As shown in Fig. 4B, only specific tryptic fragments were retained bound with GST–Rsd (compare GST–Rsd and σ70-trypsin lanes). Fragments that were recovered in the Rsd-bound fractions were then examined for their N-terminal sequences. Results indicated that GST–Rsd bound all the C terminus-proximal fragments containing region 4 of σ70. Two small proteins, R3–4 and R4, shown in Fig. 4B correspond to C-terminal fragments downstream from amino acid residues 449 and 500, respectively. The tryptic cleavage sites of σ70 were identical with those determined by Severinova et al. (28). Because the 18.5-kDa R4 fragment contains only region 4, we conclude that Rsd interacts at or downstream of the region 4 of σ70. One possibility raised by these observations is that Rsd interferes with the σ70 activity of either promoter −35 recognition or interaction with class II (σ contact) transcription factor. If this were the case, Rsd could be described as an anti-σ70 factor in stationary phase E. coli, but this key issue remains to be resolved.
Inhibition of Transcription in Vitro by Rsd Protein.
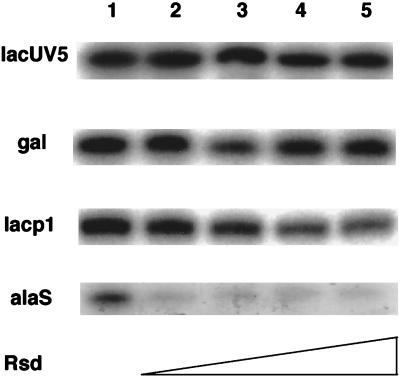
To test directly whether the Rsd protein acts as an anti-σ factor, we purified it and tested its influence on in vitro run-off transcription assays by using linear DNA templates containing σ70-regulated promoters. When lacUV5 was used as the promoter, the addition of Rsd did not produce any detectable effect on σ70 activity. On the other hand, alaS promoter-directed transcription was almost completely inhibited in the presence of excess Rsd (Fig. 5). Transcription inhibition was also measured by using other promoters. As summarized in Table 2, various extents of inhibition were observed depending on the promoter used. The effect of Rsd was also examined at increasing concentrations of templates, but all produced essentially the same effects (data not shown). When further σ70 was added, the activity was restored in a dose-dependent manner, indicating that the target of Rsd action is σ70. The variations in sensitivity to Rsd between promoters may be related to the fact that the holoenzyme concentration required to express maximum activity is not the same for all promoters. For example, the alaS promoter requires high levels of holoenzyme for maximum activity (data not shown).
Figure 5.
Effect of Rsd on in vitro transcription. A fixed amount of σ70 (1 pmol) and increasing amounts of Rsd (lanes 2–5, 1, 2, 5, and 10 pmol) were preincubated for 10 min at 30°C in our standard transcription mixture containing 50 mM NaCl (24), and then 1 pmol of core enzyme was added. After 10 min at 37°C, a DNA fragment (1 pmol) carrying the indicated promoter was added, and the mixture was incubated at 37°C for 30 min to allow the open complex formation. RNA synthesis was initiated by adding a substrate/heparin mixture and continued for 5 min at 37°C. Transcripts were analyzed by electrophoresis on 6% or 8% polyacrylamide gels containing 8 M urea.
Table 2.
Effect of Rsd on in vitro transcription
| Promoter | Transcription level, % |
|---|---|
| lacUV5 | 100 |
| galP1 | 83 |
| nusA | 83 |
| dnaQP2 | 72 |
| rnh | 72 |
| dnaQP1 | 66 |
| leuX | 62 |
| trp | 61 |
| lacP1 | 48 |
| alsS | 24 |
Single-round transcription in vitro was carried out as described in Fig. 6, using the indicated promoters (1 pmol each for both core enzyme and promoters). σ70 was preincubated for 5 min with a 10-fold molar excess of Rsd before the addition of core enzyme (core/σ70 = 1).
Intracellular Level of Rsd.
The intracellular level of Rsd was measured by using a quantitative Western blot method, as employed for measurement of the levels of various σ subunits (8, 9). Whole cell extracts were prepared from the E. coli W3110 A-type strain (18) growing at various growth phases. As shown in Fig. 6, Rsd started to increase when the rate of cell growth began to decrease. The level became maximum, reaching about 20% the level of σ70 subunit, when the cell growth stopped completely and thereafter stayed constant.
Figure 6.
Measurement of the intracellular level of Rsd. E. coli W3110 type A strain (19) was grown in LB medium at 37°C. Growth was monitored with a Klett–Summerson photometer. At the times indicated, cell lysates were prepared according to the method described previously (8, 9). The protein concentration was determined by using a protein assay kit (Bio-Rad). Aliquots containing 10 μg of total protein were subjected to the quantitative Western blot assay as employed previously (8, 9) by using the ECL reagent system (Amersham) for detection of the membrane-bound anti-Rsd antibodies.
DISCUSSION
Here, we discovered a novel stationary phase E. coli protein, Rsd, which specifically associates with σ70. Judged by N-terminal sequence analysis, all σ70-associated proteins except Rsd and ω were identified as core polymerase subunits or breakdown products of the GST–σ70 fusion protein. Several lines of evidence support the prediction that Rsd is involved in control of the activity of the σ70 subunit. (i) Rsd is formed during the growth transition from exponential to stationary phase in parallel with the shut-off of transcription of σ70-dependent genes (see Fig. 6); (ii) some of the σ70 subunit in stationary phase cell extracts exists as a complex with Rsd, which can be separated from the remaining σ70–core complexes (see Fig. 3); (iii) purified Rsd forms a specific complex in vitro with σ70 but not with other σ subunits (see Fig. 1); (iv) a binding site for Rsd on σ70 is located at or downstream from the promoter −35 binding region 4 (see Fig. 4), where some class II transcription factors also interact (3, 24); and (v) Rsd interferes with σ70-dependent transcription in vitro directed by at least some promoters (see Fig. 5 and Table 2). However, the level of transcription inhibition in vitro by Rsd was not high, presumably because: (i) the fraction of active Rsd in the overexpressed and purified Rsd fraction was low; (ii) an as yet unidentified factor(s) is involved in the formation in vivo of stable Rsd–σ70 complexes because the Rsd–σ70 complex isolated from cells was stable and was not dissociated even after isolation; or (iii) Rsd-bound σ70 loses recognition activity of only a set of promoters.
The role of anti-σ factors in the control of σ subunit activity is being increasingly recognized as a global regulatory system for transcription in prokaryotes. Regulation of the σ subunit activity by anti-σ factors is well established in Bacillus subtilis (29, 30). The existence of anti-σ factors in Salmonella typhimurium and E. coli was first identified in a regulatory system for inhibition of σF activity after completion of flagella formation (31). Synthesis of σF and FlgM (anti-σF) is induced in an early stage of the flagella cascade. The genes involved in flagella formation and chemotaxis are then transcribed by the RNA polymerase holoenzyme EσF only as long as FlgM is being excreted from the cell through immature flagella tubes. This provides a unique feedback regulation system for gene transcription, depending on the level of formation of a cellular structure. The FlgM protein is unusual because it is mostly unfolded even in the native state (and thus can be secreted through narrow flagella tubes), but it becomes structured when it binds to σF (32). The FlgM protein binds to region 4 of σF subunit and inhibits the σ function (33).
Recently a similar control of σ activity has been identified for E. coli σE, which is involved in the transcription of extreme heat-shock genes required to deal with damage to extracytoplasmic proteins (34, 35). A protein, designated RseA (Regulator of sigma E), is associated with the cell membrane and forms a complex with σE under normal growth conditions, but on exposure to certain stress conditions σE is released from the RseA complex. σE can then be used for transcription activation of the relevant genes. Thus, RseA functions as an anti-σ factor for σE. The repression of host cell gene transcription by phage T4 involves the inhibition of σ70 activity by the phage-coded, 10-kDa AsiA protein (36, 37). The AsiA protein modulates initial DNA binding by the RNA polymerase containing σ70 subunit (38). The contact site for AsiA protein on σ70, like that identified for Rsd in the present work, appears to lie near regions 3 and 4 (39). However, there is no significant similarity in overall sequence between AsiA and Rsd.
Sequence analysis of the rsd gene indicates that Rsd has a high similarity in primary structure with Pseudomonas AlgR2, a regulatory protein for exopolysaccharide alginate production (27). The AlgR2 protein is considered to regulate some enzymes involved in the pathway leading to alginate formation such as nucleoside diphosphate kinase (28), but its action mechanism remains unsolved. The identification of Rsd as an important anti-σ70 factor needs more studies in vivo, including characterization of rsd mutants.
Acknowledgments
We thank Nobuyuki Fujita for advice and Richard S. Hayward for critical reading of the manuscript. This work was supported in part by grants-in-aid from the Ministry of Education, Science and Culture of Japan and by a Core Research for Evolution Science and Technology fund from the Japan Science and Technology Cooperation.
ABBREVIATIONS
- Rsd
regulator of σD
- GST
glutathione S-transferase
- LB
Luria–Bertani
References
- 1.Helmann J D, Chamberlin M J. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 2.Gross C A, Lonetto M, Losick R. In: Transcription Regulation. McKnight S L, Yamamoto K R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 129–176. [Google Scholar]
- 3.Ishihama A. In: Mechanisms of Transcription. Eckstein F, Lilley D M J, editors. Berlin: Springer; 1997. pp. 53–70. [Google Scholar]
- 4.Ishihama A, Taketo M, Saitoh T, Fukuda R. In: RNA Polymerase. Chamberlin M, Losick R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1976. pp. 475–502. [Google Scholar]
- 5.Ishihama A. In: Control of Cell Growth and Division. Ishihama A, Yoshikawa H, editors. Heidelberg: Springer; 1991. pp. 121–140. [Google Scholar]
- 6.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Mau B, Shao Y. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Iwakura Y, Ishihama A. Mol Gen Genet. 1974;142:1–23. doi: 10.1007/BF00268673. [DOI] [PubMed] [Google Scholar]
- 8.Jishage M, Ishihama A. J Bacteriol. 1995;177:6832–6835. doi: 10.1128/jb.177.23.6832-6835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jishage M, Iwata A, Ueda S, Ishihama A. J Bacteriol. 1996;178:5447–5451. doi: 10.1128/jb.178.18.5447-5451.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saitoh T, Ishihama A. J Mol Biol. 1977;115:403–416. doi: 10.1016/0022-2836(77)90162-0. [DOI] [PubMed] [Google Scholar]
- 11.Mulvey M R, Loewen P C. Nucleic Acids Res. 1989;17:9979–9991. doi: 10.1093/nar/17.23.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange R N, Hengge-Aronis R. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka K, Takayanagi Y, Fujita N, Ishihama A, Takahashi H. Proc Natl Acad Sci USA. 1993;90:3511–3515. doi: 10.1073/pnas.90.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loewen P C, Hengge-Aronis R. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 15.Muffler A, Traulsen D D, Lange R, Hengge-Aronis R. J Bacteriol. 1996;178:1607–1613. doi: 10.1128/jb.178.6.1607-1613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishihama A. Curr Opin Genet Dev. 1997;7:582–588. doi: 10.1016/s0959-437x(97)80003-2. [DOI] [PubMed] [Google Scholar]
- 17.Blattner F R, Burland V, Plunkett G, III, Sofia H J, Daniels D. Nucleic Acids Res. 1993;21:5408–5417. doi: 10.1093/nar/21.23.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jishage M, Ishihama A. J Bacteriol. 1997;179:959–963. doi: 10.1128/jb.179.3.959-963.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Igarashi K, Ishihama A. Cell. 1991;65:1015–1022. doi: 10.1016/0092-8674(91)90553-b. [DOI] [PubMed] [Google Scholar]
- 20.Ding Q, Kusano S, Villarejo M, Ishihama A. Mol Microbiol. 1995;16:649–656. doi: 10.1111/j.1365-2958.1995.tb02427.x. [DOI] [PubMed] [Google Scholar]
- 21.Kundu T K, Kusano S, Ishihama A. J Bacteriol. 1997;179:4264–4269. doi: 10.1128/jb.179.13.4264-4269.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kusano S, Ding Q, Fujita N, Ishihama A. J Biol Chem. 1996;271:1998–2004. doi: 10.1074/jbc.271.4.1998. [DOI] [PubMed] [Google Scholar]
- 23.Kajitani M, Ishihama A. Nucleic Acids Res. 1983;11:671–686. doi: 10.1093/nar/11.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar A, Grimes B, Fujita N, Makino K, Malloch R A, Hayward R S, Ishihama A. J Mol Biol. 1994;235:405–413. doi: 10.1006/jmbi.1994.1001. [DOI] [PubMed] [Google Scholar]
- 25.Nomura T, Fujita N, Ishihama A. Nucleic Acids Res. 1986;14:6857–6870. doi: 10.1093/nar/14.17.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato J, Chu L, Kitano K, De Vault J D, Kimbara K, Chakrabarty A M, Misra T K. Gene. 1989;84:31–38. doi: 10.1016/0378-1119(89)90136-4. [DOI] [PubMed] [Google Scholar]
- 27.Schlictman D, Kubo M, Shankar S, Chakrabarty A M. J Bacteriol. 1995;177:2469–2474. doi: 10.1128/jb.177.9.2469-2474.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Severinova E, Severinov K, Fenyo D, Marr M, Brody E N, Roberts J W, Chait B T, Darst S A. J Mol Biol. 1996;263:637–647. doi: 10.1006/jmbi.1996.0604. [DOI] [PubMed] [Google Scholar]
- 29.Brown K L, Hughes K T. Mol Microbiol. 1995;16:397–404. doi: 10.1111/j.1365-2958.1995.tb02405.x. [DOI] [PubMed] [Google Scholar]
- 30.Stragier P, Losick R. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 31.Ohnishi K, Kutsukake K, Suzuki H, Iino T. Mol Microbiol. 1992;6:3149–3157. doi: 10.1111/j.1365-2958.1992.tb01771.x. [DOI] [PubMed] [Google Scholar]
- 32.Daughdrill G W, Chadsey M S, Karlinsey J E, Hughes K T, Dahlquist F W. Nat Struct Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- 33.Kutsukake K, Iyoda S, Ohnishi K, Iino T. EMBO J. 1994;13:4568–4576. doi: 10.1002/j.1460-2075.1994.tb06778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Missiakas D, Mayer M P, Marc L, Georgopoulus C, Raina S. Mol Microbiol. 1997;24:355–371. doi: 10.1046/j.1365-2958.1997.3601713.x. [DOI] [PubMed] [Google Scholar]
- 35.De Las Penas A, Connolly L, Gross C A. Mol Microbiol. 1997;24:373–385. doi: 10.1046/j.1365-2958.1997.3611718.x. [DOI] [PubMed] [Google Scholar]
- 36.Brody E N, Kassavetis G A, Ouhammouch M, Sanders G M, Tinker R L, Geiduschek E P. FEMS Lett. 1995;128:1–8. doi: 10.1111/j.1574-6968.1995.tb07491.x. [DOI] [PubMed] [Google Scholar]
- 37.Orshini G, Ouhammouch M, Lecaer J P, Brody E N. J Bacteriol. 1993;175:85–93. doi: 10.1128/jb.175.1.85-93.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adelman K, Orsini G, Kolb A, Graziani L, Brody E N. J Biol Chem. 1997;272:27435–27443. doi: 10.1074/jbc.272.43.27435. [DOI] [PubMed] [Google Scholar]
- 39.Hinton D M, March-Amegadzie R, Gerber J S, Sharma M. J Mol Biol. 1996;256:235–248. doi: 10.1006/jmbi.1996.0082. [DOI] [PubMed] [Google Scholar]