Abstract
The hetR gene plays a very important role in cell differentiation of heterocystous cyanobacteria. To understand the mechanism of the hetR gene product in regulation of heterocyst differentiation, the recombinant HetR protein (rHetR) was overproduced in Escherichia coli. Purified rHetR was unstable and degraded easily in solution. Phenylmethanesulfonyl fluoride, a serine-type protease inhibitor, prevented the degradation and was shown to modify covalently rHetR. Dansyl fluoride (DnsF), another serine-type protease inhibitor, also covalently modifies rHetR as shown by electrophoresis and electroblotting of the labeled rHetR and by MS. The labeling of rHetR with phenylmethanesulfonyl fluoride and DnsF was at the same site of rHetR and required Ca2+. S179N-rHetR, a mutant protein from strain 216 of Anabaena PCC 7120, which cannot differentiate heterocysts because of the mutation, was also overproduced and characterized. Although S170N-rHetR still can be labeled with DnsF, no proteolysis was observed, suggesting that Ser179 is involved in proteolytic activity. DnsF-labeled rHetR was digested with trypsin, and the labeled peptide was isolated and sequenced. The labeled peptide matches a sequence from HetR. These results show that HetR is a protease.
Keywords: cyanobacteria, Anabaena PCC7120, heterocyst differentiation
Cyanobacteria are a diverse group of prokaryotes that carry out oxygenic photosynthesis. Some cyanobacteria also can fix dinitrogen. The two processes are separated either temporally or spatially because oxygen is detrimental to nitrogenase. Some filamentous cyanobacteria that perform nitrogen fixation have specialized cells called heterocysts where nitrogenase is located (1–4). Heterocysts are formed when combined nitrogen in the growth medium is depleted and when the number of vegetative cells between two existing heterocysts on a filament is large enough. In the process of differentiation from a vegetative cell to a heterocyst, many morphological and biochemical changes occur, and most of them are regulated at the level of gene expression (2, 3). In most of the filaments, heterocysts are spaced regularly so that there is a pattern along the filaments.
The hetR gene from Anabaena PCC 7120 first was reported by Buikema and Haselkorn (5). They showed that it absolutely was required for heterocyst differentiation and that pattern formation also was influenced strongly by the expression of this gene. Shifting from a nitrogen-replete condition to a nitrogen-depleted condition resulted in up-regulation of hetR gene transcription, and the transcripts of the hetR gene were present mostly in those cells that would become heterocysts and proheterocysts (6). The up-regulation of the hetR gene transcription requires the presence of a functional hetR gene product, suggesting that the hetR gene is under the control of positive feedback (6). The hetR gene is also critical to akinete formation (7) and may be required in other cellular processes in nonfilamentous cyanobacteria (5).
Little is known about the mechanism by which the hetR gene product regulates cell differentiation. The deduced amino acid sequence shows no similarity to any other protein, and no apparent DNA binding motif was observed. We recently have succeeded in overproducing recombinant HetR protein and have raised antibodies against rHetR. Immunoblotting results showed that the metabolism of the HetR protein in vivo was related closely to the process of heterocyst differentiation (8). In this report, we describe crystallization and biochemical characterization of the HetR protein. Our results show that HetR could function as a protease in heterocystous cyanobacteria.
MATERIALS AND METHODS
Recombinant HetR Protein.
The coding sequence of the wild-type hetR gene of Anabaena PCC 7120 and the mutant hetR gene encoding a Ser179Asn mutation (S179N) from strain 216 of Anabaena PCC 7120 (a kind gift from Robert Haselkorn and William J.Buikema, University of Chicago) (5) were amplified by PCR and cloned into pET-3a (9). The PCR was carried out with pfu DNA polymerase in addition to Taq DNA polymerase for high fidelity (10). The resultant expression plasmids pET3a-hetR and pET3a-hetRm containing the wild-type hetR gene and the mutant hetR gene, respectively, were transformed into Escherichia coli strain BL21(DE3). Overproduction of the recombinant HetR protein (rHetR) and S179N-rHetR was achieved by induction with isopropyl-β-d-thiogalactopyranoside. Isolation of rHetR inclusion bodies and refolding of rHetR in solution were carried out according to Zhao et al. (11) except that urea was replaced with 6 M guanidine HCl. The refolded rHetR and S179N-rHetR were purified to homogeneity with DEAE-Sephadex and Sephacel S-200 chromatography. The rHetR then was concentrated to ≈10 mg/ml by using ultra-filtration through a 10-kDa cut-off membrane (Amicon).
Crystals of rHetR were grown with the standard vapor diffusion method from a protein solution of 10 mg/ml in 1 M NaCl. The protein solution was put into a 0.5-ml centrifuge tube with the lid removed, and the tube was inserted into a 1.5-ml tube containing various concentrations of NaCl. The 1.5-ml tube then was sealed and was left at 4°C for 1 week. The crystals that formed were transferred to a microscope slide, were maintained hydrated with a solution of 25% (wt/vol) polyethylene glycol 6000, and were photographed with a Leica (Deerfield, IL) microscope equipped with a camera.
Degradation of rHetR and S179N-rHetR was studied as follows. The rHetR and S179N-rHetR at a concentration of 1 mg/ml in TS buffer (20 mM Tris⋅Cl, pH7.5/100 mM NaCl) containing 1 mM CaCl2 was incubated for various times at 37°C with or without a prior 95°C treatment for 2 min. Phenylmethanesulfonyl fluoride (PMSF) was added to a final concentration of 100 μM when needed. After the incubation, the protein solutions were subjected to SDS/PAGE according to Laemmli (12). The protein and its fragments were visualized by silver staining (13).
Protease Inhibitor Reaction with rHetR.
Reaction of rHetR and S179N-rHetR with PMSF and 5-dimethylaminonaphthalene-1-sulfonyl fluoride, or dansyl fluoride (DnsF), was performed according to Vaz and Schoellmann (14) either in the presence of CaCl2 at concentrations indicated in the text or in the presence of EGTA at a concentration of 1 mM. The PMSF-modified rHetR was analyzed by nondenaturing gel electrophoresis according to Laemmli (12) except that SDS was omitted from the loading buffer and the gel was run at 4°C. The gel then was stained with Coomassie brilliant blue. The rHetR reacted with DnsF first was precipitated with 80% acetone to remove unincorporated DnsF and then was subjected to either reducing (in the presence of 10 mM DTT) or nonreducing (without DTT) SDS/PAGE, followed by electrophoretic transfer to a polyvinylidene fluoride (PVDF) membrane (ProBlott, Applied Biosystems). The fluorescent bands were recorded with a digital camera (Bio-Rad). The membrane then was stained with Coomassie brilliant blue to visualize the protein bands.
To identify the DnsF-labeled tryptic fragment, the fluorescent band on the PVDF membrane was cut out, and the protein was digested with trypsin according to Fernandez et al. (15). The membrane then was extracted twice with 50% acetonitrile in 1% trifluoroacetic acid and concentrated under vacuum to reduce the volume to ≈5 μl. The solution then was injected to a micro-HPLC (ABI173, Applied Biosystems). The elution gradient was 0–45% acetonitrile in 0.1% trifluoroacetic acid at a flow rate of 10 μl/min. The eluted peptides were blotted directly onto a PVDF strip by using a Dynamic Blotter system equipped with the HPLC. The fluorescent spot was visualized under long wavelength UV light and was cut out for peptide sequencing.
Other Methods.
E. coli strains were grown in Luria–Bertani medium with 50 μg/ml ampicillin when needed. DNA was sequenced with an ABI377 automatic DNA sequencer (Applied Biosystems), and N-terminal protein sequencing was performed with an ABI491 protein sequencer (Applied Biosystems) at the core lab facilities of the College of Life Sciences, Peking University, Beijing. Protein was loaded directly onto a piece of PVDF before sequencing. Sequencing of proteins on PVDF membranes was carried out in a gas phase sequencing procedure (16). MS determination of the molecular masses of rHetR was performed with a G2025A MS instrument (Hewlett Packard). The matrix used in matrix-assisted laser desorption time-of-flight was sinapinic acid. Protein concentration was determined with Coomassie brilliant blue (17). All chemicals were purchased from Sigma unless otherwise stated. All enzymes used in molecular cloning were purchased from Promega.
RESULTS
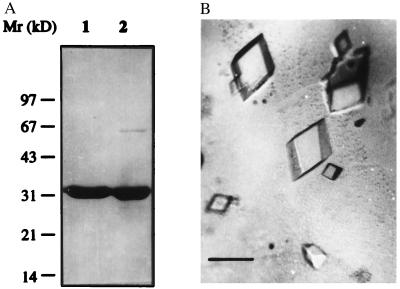
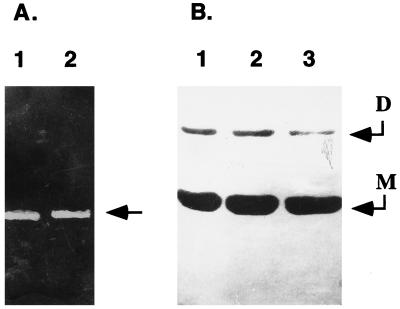
Fig. 1A shows SDS/PAGE of the purified recombinant HetR protein (rHetR) under reducing (lane 1) and nonreducing conditions (lane 2). The apparent molecular mass of rHetR was ≈33 kDa. Under nonreducing conditions, there was an additional weak band that had a molecular mass of 66 kDa, corresponding to a dimer of rHetR. N-terminal protein sequencing confirmed that both bands were rHetR with the initial methionine residue removed (data not shown). Fig. 1B shows rHetR crystals generated with NaCl as precipitant. Although we were able to generate crystals from HetR with ammonium sulfate and polyethylene glycol 6000, NaCl always gave the best results. The large crystals shown in Fig. 1 have a rhombohedral form with average dimensions of 0.5 × 0.5 × 0.25 mm. We replaced urea with guanidine HCl in the refolding process because the latter solubilized rHetR inclusion bodies better and more crystals could be generated. We currently are collecting x-ray diffraction data from the crystals.
Figure 1.
Crystallization of rHetR. (A) SDS/PAGE of purified rHetR under reducing (lane 1) and nonreducing (lane 2) conditions. The purified rHetR was in TS (see text) buffer containing EGTA (1 mM) after the final step of purification. The molecular mass standards (in kilodaltons) are shown on the left. (B) Photographs of rHetR crystals. Conditions for crystal formation with NaCl as precipitant are described in Materials and Methods. The crystals are mounted on a slide in polyethylene glycol 6000 solution and photographed through a Leica microscope. The average sizes of large crystals are 0.5 × 0.5 × 0.25 mm. (Bars = 0.5 mm.)
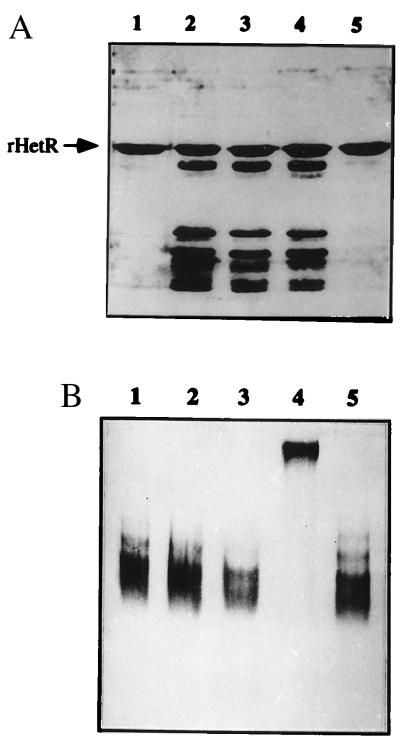
In the process of crystal generation, we observed that rHetR was degraded easily in solution even though our preparation of rHetR was apparently pure. We investigated the degradation of rHetR without addition of any proteases with the results shown in Fig. 2A. Incubation of the purified rHetR at 37°C resulted in a pattern of degradation fragments (lanes 2 through 4). All of the fragments generated were from rHetR as confirmed by immunoblotting (data not shown). The degradation of rHetR in vitro could be prevented by high temperature incubation for 2 min before 37°C treatment (Fig. 2A, lane 1) or by the addition of a serine-type protease inhibitor, PMSF (Fig. 2A, lane 5). These results indicated that there was activity of a serine-type protease in the rHetR solution. This led us to study the possibility that HetR is a protease, as suggested by Buikema and Haselkorn (2, 5). The rHetR was incubated with PMSF under various conditions for 1 hr and run in a nondenaturing gel (Fig. 2B). We found that the mobility of HetR was much lower when Ca2+ (10 μM) and PMSF (100 μM) were both present (Fig. 2B, lane 4). Incubation with PMSF (Fig. 2B, lane 2), Ca2+ (Fig. 2B, lane 3), or only with isopropanol (0.2%, vol/vol), which was used to dissolve PMSF (Fig. 2B, lane 5), had no effect on rHetR’s mobility in the nondenaturing gel electrophoresis as compared with the control (Fig. 2B, lane 1).
Figure 2.
(A) Degradation of rHetR in vitro. Purified rHetR solution at 1 mg/ml was incubated in TS buffer containing 1 mM CaCl2 for 1 h (lane 2), 2 h (lane 3), and 4 h (lane 4) at 37°C. In lane 5, rHetR was treated as in lane 2 except that PMSF was present at a concentration of 100 μM. Lane 1 contained rHetR that was heated at 95°C for 2 min before the incubation. All samples were treated with sample loading buffer before SDS/PAGE. The gel was silver-stained. (B) PMSF effect on the mobility of rHetR in nondenaturing gel electrophoresis. The rHetR was incubated for 1 h at room temperature in TS buffer without addition (lane 1), with 1 mM EGTA and 100 μM PMSF (lane 2), with 10 μM CaCl2 (lane 3), with 100 μM PMSF and 10 μM CaCl2 (lane 4), and with 0.2% isopropanol only (lane 5). The treated samples were analyzed by nondenaturing gel electrophoresis and stained with Coomassie brilliant blue.
We further investigated the specific labeling of rHetR with another serine-type protease inhibitor, DnsF. DnsF reacts with the active serine of serine-type proteases specifically (14). It has an additional advantage that the labeled adduct is fluorescent and can be identified easily. The rHetR was incubated with DnsF under different conditions overnight at 4°C, and the protein was precipitated with 80% acetone to remove free DnsF. The labeled protein then was solubilized and subjected to SDS/PAGE. Because free acrylamide is a very effective quencher of the DnsF fluorescence (18), we transferred the protein in gel to a PVDF membrane, and the fluorescent bands were photographed under UV light (Fig. 3). Incubation of rHetR with DnsF in the presence of 10 μM Ca2+ resulted in a fluorescent adduct (Fig. 3A, lane 3). Because the SDS/PAGE was performed under nonreducing condition, the dimer form was also present, and it was fluorescent. Incubation with PMSF and DnsF together resulted in a reduced intensity of the bands (Fig. 3A, lane 2), suggesting that both inhibitors compete for the same site on rHetR. When Ca2+ was omitted from the incubation buffer and EGTA was added, much less fluorescence could be observed in the rHetR position (Fig. 3A, lane 4). Fig. 3B was the same PVDF membrane shown in Fig. 3A except that it was stained with Coomassie blue to show the relative amount of rHetR in each lane.
Figure 3.
Covalent modification of rHetR with DnsF. (A) The rHetR (1 mg/ml) was incubated with 1 mM DnsF in the presence of 10 μM CaCl2 and 100 μM PMSF (lane 2), 10 μM CaCl2 alone (lane 3), or 1 mM EGTA without added Ca2+ (lane 4). Lane 1 contains proteins of molecular mass standards incubated with DnsF as in lane 3. The samples were precipitated with 80% acetone before SDS/PAGE under nonreducing conditions. The proteins in the gel then were electroblotted to a ProBlott (PVDF) membrane. The membrane was soaked in pure acetone for 10 min to remove any free DnsF. The fluorescent bands were recorded with a digital camera. (B) The same membrane shown in Fig. 3A except that it was stained with Coomassie brilliant blue.
In the process of the DnsF labeling, we observed several times more dimer form when Ca2+ was present. Fluorescent dimer formation showed that the binding site for DnsF was not the single Cys residue. The fluorescent band also was sequenced at the N terminus, confirming that the band was indeed from rHetR (not shown). The accessibility of the N terminus of the labeled rHetR showed that DnsF, unlike dansyl chloride, does not block the protein’s amino terminal and indicated that the labeling was specific as in other cases studied with DnsF (14).
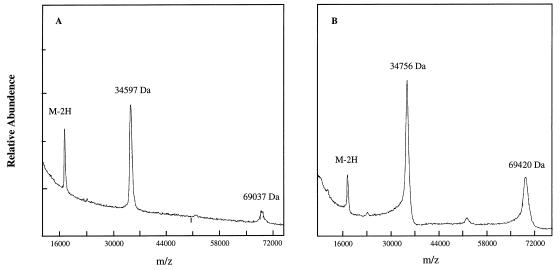
To confirm further that rHetR was labeled specifically with DnsF, time-of-flight mass spectra were obtained to determine the molecular mass of the rHetR–DnsF adduct with the results shown in Fig. 4. The monomer and dimer of wild-type rHetR had average molecular masses of 34,597 Da and 69,037 Da, respectively, very close to those of the calculated values (Fig. 4A). The monomer and dimer of the labeled rHetR had average molecular masses of 34,756 Da and 69,420 Da, respectively. The labeled monomer was 159 Da larger than the rHetR, and the labeled dimer was 383 Da larger than the dimer before labeling (Fig. 4B). If all of the rHetR was labeled with DnsF, it would have a molecular mass increase of 234 Da for the monomer and 468 Da for the dimer. The smaller values of molecular mass increase after labeling suggested that the labeling was not complete as in cases of studying proteases with DnsF (14). The MS method of time-of-flight used in our study could only give an average molecular mass of both labeled and unlabeled forms. The values obtained suggested that >67% of the monomer rHetR and >81% of the dimer rHetR were labeled, respectively. The mass spectra also show that more dimer forms were present in the presence of Ca2+.
Figure 4.
Matrix-assisted laser desorption time-of-flight of unlabeled (A) and dansyl-labeled (B) rHetR. HetR samples (≈100 pmol) were treated as described in Materials and Methods and analyzed with a G2025A MS instrument. The molecular masses of the peaks detected are shown in each spectrum.
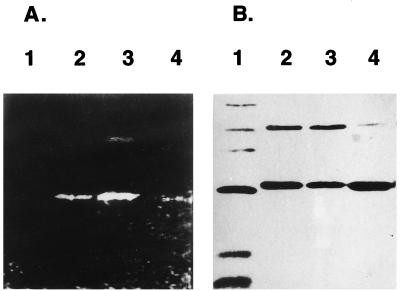
The discovery of the hetR gene from Anabaena PCC 7120 was based on a serine to asparagine mutation at position 179, which resulted in a nonfunctional gene (5). So the first choice to identify the active serine that reacted with DnsF was to study the serine at position 179. The mutant gene encoding S179N-rHetR was amplified from strain 216 (5) and confirmed by DNA sequencing (not shown). It then was overexpressed in E. coli. The gene product was refolded and was purified as rHetR. When the purified S179N-rHetR was incubated with DnsF in the presence of Ca2+, we found that it also became fluorescent with approximately the same intensity (Fig. 5A, lane 2) as compared with rHetR (lane 1). It was apparent that the Ser179 was not required for labeling with DnsF in vitro even though it absolutely was required for HetR normal function in vivo.
Figure 5.
Characterization of S179N-rHetR. (A) DnsF labeling of S179N-rHetR. Wild-type rHetR and S179N-rHetR were labeled with DnsF, and the fluorescent bands after SDS/PAGE and electroblotting were recorded as described in Fig. 3. (B) Lack of proteolytic activity of S179N-rHetR. S179N-rHetR at a concentration of 1 mg/ml was incubated at 37°C for 1 h (lane 2) or 4 h (lane 3) or was not incubated at all before SDS/PAGE in the absence of DTT. The incubation condition was as described in Fig. 2. The gel was silver-stained. The dimer and monomer are indicated by arrows.
To further investigate Ser179 function, in vitro proteolysis of S179N-rHetR was studied with the results shown in Fig. 5B. Incubation of S179N-rHetR at 37°C for 1 h (Fig. 5B, lane 2) and 4 h (Fig. 5B, lane 3) in the presence of Ca2+ did not yield any proteolytic fragments as visualized with silver staining. The band intensities were the same as that of the control (Fig. 5B, lane 1). These results suggest that rHetR is autodigestive and that Ser179 is required for proteolytic activity. We again observed that a dimer of S179N-rHetR formed in the presence of calcium (Fig. 5B).
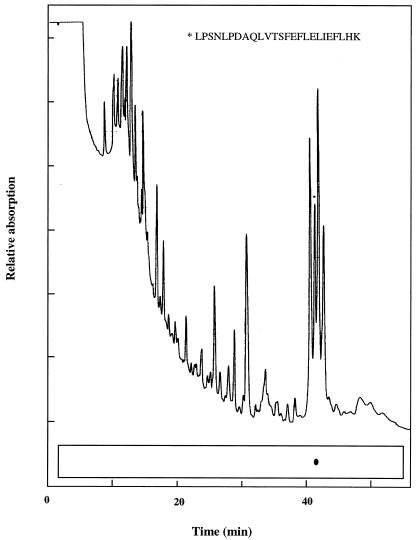
To determine the actual site of DnsF modification, a tryptic peptide containing the dansyl group was isolated from DnsF-modified rHetR (Fig. 6). We chose to digest the electroblotted fluorescent rHetR band with trypsin in situ because this essentially would eliminate any interference of free DnsF with trypsin. This procedure generated only one fluorescent spot under UV light after blotting the peptides onto a strip of PVDF membrane. The spot was cut out and sequenced from its amino terminus. The sequencing result was LPSNLPDAQLVTSFEFLELIEFLHK. This sequence matched the HetR peptide from position 140 to 164. The peptide was preceded by a lysine residue and terminated with a lysine residue, showing that it is a tryptic peptide. There are two serine residues on this tryptic fragment, and both of them could be identified positively at the standard phenylthiohydantoin–Ser position in peptide sequencing. It has been reported that the covalent bond between the dansyl group and the hydroxyl group is not stable in a strongly acidic environment (14) such as when trifluoroacetic acid is present in peptide sequencing. Mutations at these two serine positions currently are being introduced to determine which serine residue is the binding site of the dansyl group.
Figure 6.
Peptide mapping of the dansyl-labeled rHetR. The labeled rHetR was run on SDS/PAGE and electroblotted onto a PVDF membrane as in Fig. 3. The fluorescent band was cut into small pieces and treated with trypsin at 5 μg/ml overnight at 37°C. The total digestion volume was 25 μl. The digested rHetR was eluted from the PVDF membrane with 50% acetonitrile in 1% trifluoroacetic acid, and the tryptic peptides were separated with a micro-HPLC. The separated peptides were blotted directly onto a strip of PVDF membrane, and the fluorescent spot was visualized under long UV light (schematically shown here as a dot) and cut out for peptide sequencing. The peak corresponding to the fluorescent spot is marked with an asterisk. The sequence of the peptide in the marked peak is shown.
DISCUSSION
The ability to generate crystals of rHetR strongly suggests that the refolded rHetR is in a homogeneous conformation, which is critical to biochemical studies of rHetR. The rHetR crystals reported here were large enough for x-ray diffraction study of its three dimensional structure. Degradation of wild-type rHetR was demonstrated readily by incubation of purified rHetR at 37°C for 1 hr. It can be prevented by PMSF or by heating the protein solution at 95°C (Fig. 2). No degradation of S179N-rHetR was observed under the same conditions (Fig. 5). These results suggest that degradation of rHetR resulted from rHetR itself, not from contaminant protease in the rHetR preparation because S179N-rHetR was prepared in an identical way. Our results also show that Ser179 is a key residue required for rHetR autodegradation.
Because PMSF has been shown to be a specific serine-type protease inhibitor, demonstration of covalent modification of rHetR with PMSF would be strong evidence that HetR is a protease. In the study of PMSF reaction with rHetR, we used a nondenaturing gel system based on the assumption that a covalently modified rHetR with PMSF would have more negative charge and would move faster than the unmodified rHetR. Even though our results showed that incubation of rHetR with PMSF in the presence of Ca2+ resulted in a slower mobility, this result does suggest that rHetR was modified covalently by PMSF and that the modification by PMSF required Ca2+. The reason for the low mobility of the PMSF-modified rHetR in the nondenaturing gel system is unknown. We found no previous report that proteases behave similarly. We did find in a preliminary test that PMSF-modified trypsin ran more slowly in the nondenaturing gel system in the presence of Ca2+ (R.Z. and J.Z., unpublished result). DnsF has been shown to be a highly specific serine-type protease inhibitor, reacting with a serine residue at the active site (14). It has been used to study structure and function of proteases such as subtilisins (18) and chymotrypsin (14). Incubation of rHetR with DnsF indeed resulted in a fluorescent adduct, and this reaction again was dependent on the presence of Ca2+. The specific labeling with DnsF was confirmed by matrix-assisted laser desorption time-of-flight. PMSF competed with DnsF for the active site (Fig. 3), suggesting that the two inhibitors reacted with the same site. The evidence that rHetR is autodigestive and that specific labeling of rHetR with both PMSF and DnsF strongly suggests that HetR is a serine-type protease. It is apparent that HetR is not a typical serine-type protease because no similarity between HetR and other serine-type proteases was found. HetR may belong to a serine-type protease group with an unusual structure, such as a protease found in the fungus Conidiobolus coronatus (19).
The fact that the protease inhibitor reaction with rHetR requires Ca2+ suggests that the activity of HetR in vivo depends on Ca2+. Ca2+-dependent protease activities were reported in the process of cell differentiation of Anabaena PCC 7120 (20). Maldener et al. (21) reported cloning and sequencing a Ca2+-dependent protease gene, prcA, from Anabaena variabilis (21) although its importance in cell differentiation is still unclear. Ca2+-dependent proteases also are present in other organisms, such as subtilisins in Bacillus (22).
The dimer formation of rHetR through a covalent disulfide bond was confirmed with SDS/PAGE under nonreducing conditions and with MS. The physiological significance of the dimer formation is unknown. Although serine-type protease inhibitors such as PMSF have been shown to be inhibitory to some cysteine-type proteases, the formation of a dansyl-labeled dimer rHetR essentially eliminated the possibility that the active site is a cysteine residue because there is only one cysteine residue per HetR protein.
Heterocyst differentiation is a complex process. One of the early events in this process is the up-regulation of the hetR gene (5), which depends on a correct gene product of its own (6). The hetR gene was suggested to be the master switch controlling cell differentiation (3) and pattern formation (5). The mechanism of the HetR protein in regulation of cell differentiation is not understood. It was suggested that HetR could be a protease (5) based on the fact that S179N mutation led to a total loss of its function. Although peptide mapping and sequencing results showed that the ansyl group was not attached to Ser179, our results show that Ser179 is required for the protease activity. Several proteins, such as bovine serum albumin, were incubated with rHetR, and no obvious proteolysis was found (data not shown). We know little about the substrates of HetR except that rHetR can digest itself (Fig. 2). The Ser179 of HetR could be involved in substrate binding. It is even possible that the dansyl-labeled serine residue is just within the pocket of the active site whereas the real active serine participating in enzymatic digestion is the Ser179. We recently have found that the pI of the native HetR under nitrogen-deplete conditions is much more acidic than that of rHetR (8). One possibility is that HetR in vivo is autodigestive until it is modified after cells receive the signal of nitrogen deprivation.
In many organisms, proteases play important roles in cell differentiation and development. Caspases are proteases required for normal development of animals such as the Pcd-1 in Drosophila (23) and Ced-3 in nematodes (24) although little is known about their mechanism. Cell cycles also are influenced strongly by protease activities (25). In Bacillus subtilis, a serine-type protease is required during sporulation (26). It is conceivable that HetR functions as a protease that digests repressors of the genes to be turned on and activators of the genes to be turned off under nitrogen deprivation conditions. During the early phases of heterocyst differentiation, intracellular proteolysis activity increased several times in Anabaena (27, 28). Many proteins of vegetative cells are degraded by a Ca2+-requiring protease activity in the differentiation process (20). Even though the strains with mutated prcA genes showed no signs of impairment of heterocyst differentiation, the activity of the PrcA may not include necessarily all of the Ca2+-dependent protease activities in vivo. HetR could provide the driving activity for cell differentiation as in the case of protease activities controlling cell cycles of budding yeast (25).
Acknowledgments
We thank Professors X. Wu, G. Wu, and Z. Cao for their suggestions. We also thank Mr. X. Li for technical assistance. This work was supported by National Natural Science Foundation of China to J. Z. (grants 39535002 and 39570067).
ABBREVIATIONS
- DnsF
5-dimethylaminonaphthalene-1-sulfonyl fluoride, or dansyl fluoride
- PMSF
phenylmethanesulfonyl fluoride
- PVDF
polyvinylidene fluoride
- rHetR
recombinant HetR protein
References
- 1.Haselkorn R. Annu Rev Plant Physiol. 1978;29:319–344. [Google Scholar]
- 2.Buikema W J, Haselkorn R. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:33–52. [Google Scholar]
- 3.Wolk C P, Ernst A, Elhai J. In: Molecular Biology of the Cyanobacteria. Bryant D A, editor. Dordrecht, The Netherlands: Kluwer; 1995. pp. 769–823. [Google Scholar]
- 4.Fay P. Microbiol Rev. 1992;56:340–373. doi: 10.1128/mr.56.2.340-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buikema W J, Haselkorn R. Genes Devel. 1991;5:321–330. doi: 10.1101/gad.5.2.321. [DOI] [PubMed] [Google Scholar]
- 6.Black T A, Cai Y, Wolk C P. Mol Microbiol. 1993;9:77–84. doi: 10.1111/j.1365-2958.1993.tb01670.x. [DOI] [PubMed] [Google Scholar]
- 7.Leganes F, Fernandez–Pinas F, Wolk C P. Mol Microbiol. 1994;12:679–684. doi: 10.1111/j.1365-2958.1994.tb01055.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhou, R., Cao, Z. & Zhao, J. (1998) Arch. Microbiol., in press. [DOI] [PubMed]
- 9.Studier F W, Rosenberg A H, Dunn J J, Dubenforff J W. Methods Enzymol. 1991;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 10.Barnes W M. Proc Natl Acad Sci USA. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, Li N, Warren P V, Golbeck J H, Bryant D A. Biochemistry. 1992;31:5093–5099. doi: 10.1021/bi00137a001. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli U K. Nature (London) 1971;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Andrews A T. Electrophoresis. Oxford: Oxford Univ. Press; 1992. [Google Scholar]
- 14.Vaz W L C, Schoellmann G. Biochem Biophys Acta. 1976;439:194–205. doi: 10.1016/0005-2795(76)90175-6. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez J, DeMott M, Atherton D, Mische S M. Anal Biochem. 1992;201:255–264. doi: 10.1016/0003-2697(92)90336-6. [DOI] [PubMed] [Google Scholar]
- 16.Matsudaira P. J Biol Chem. 1987;262:10035–10039. [PubMed] [Google Scholar]
- 17.Branford D, Chramback A. Anal Biochem. 1971;40:95–134. doi: 10.1016/0003-2697(71)90086-8. [DOI] [PubMed] [Google Scholar]
- 18.Genov N C, Boteva R N. Biochem J. 1986;238:923–926. doi: 10.1042/bj2380923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phadtare S U, Rao M, Vasanti D. Arch Microbiol. 1997;166:414–417. doi: 10.1007/BF01682989. [DOI] [PubMed] [Google Scholar]
- 20.Wood N B, Haselkorn R. In: Limited Proteolysis in Microorganisms. Cohen G N, Holzer H, editors. Education, and Welfare, Washington, D.C.: U. S. Department of Health; 1979. Publ. No. (NIH) 79–1591, pp. 159–166. [Google Scholar]
- 21.Maldener I, Lockau W, Cai Y, Wolk C P. Mol Gen Genet. 1991;225:113–120. doi: 10.1007/BF00282649. [DOI] [PubMed] [Google Scholar]
- 22.Serrano L, Avila J, Maccioni R B. Biochemistry. 1984;23:4675–4681. doi: 10.1021/bi00315a024. [DOI] [PubMed] [Google Scholar]
- 23.Song Z, McCall K, Steller H. Science. 1997;275:536–540. doi: 10.1126/science.275.5299.536. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S. Trends Biochem Sci. 1995;20:198–199. doi: 10.1016/s0968-0004(00)89007-6. [DOI] [PubMed] [Google Scholar]
- 25.King R W, Deshaies R J, Peter J, Kirschner M W. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 26.Dancer B N, Mandelstam J. J Bacteriol. 1975;121:406–410. doi: 10.1128/jb.121.2.406-410.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ownby J D, Shannahan M, Hood E. J Gen Microbiol. 1979;110:255–261. [Google Scholar]
- 28.Wood N B, Haselkorn R. J Bacteriol. 1980;141:1375–1385. doi: 10.1128/jb.141.3.1375-1385.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]