Abstract
The thermodynamic stabilities of small protein domains are clearly subject to natural selection, but it is less clear whether the rapid folding rates typically observed for such proteins are consequences of direct evolutionary optimization or reflect intrinsic physical properties of the polypeptide chain. This issue can be investigated by comparing the folding rates of laboratory-generated protein sequences to those of naturally occurring sequences provided that the method by which the sequences are generated has no kinetic bias. Herein we report the folding thermodynamics and kinetics of 12 heavily mutated variants of the small IgG binding domain of protein L retrieved from high-complexity combinatorial libraries by using a phage-display selection for proper folding that does not discriminate between rapidly and slowly folding proteins. Although the stabilities of all variants were decreased, many of the variants fold faster than wild type. Taken together with similar results for the src homology 3 domain, this observation suggests that the sequences of small proteins have not been extensively optimized for rapid folding; instead, rapid folding appears to be a consequence of selection for stability.
For proteins to fold successfully, their native states must be stable and kinetically accessible. Protein stability is clearly under selective pressure because the native states of most proteins must be significantly populated for proper protein function. There are reasons to suspect that folding rates may also be under selective pressure: protein folding time scales of milliseconds to minutes are many orders of magnitude slower than the time for an exhaustive search of conformational space (1) and are biologically convenient because slower folding rates would make folding the rate-limiting step in protein synthesis and could compromise cell viability. Furthermore, there are very large free-energy barriers in some folding reactions; the native states of a number of proteases are not accessible under biological time scales in the absence of accessory factors (2, 3). Finally, recent experiments have suggested that specific packing interactions in a small protein (CI2) may be optimized for the rate of folding at the expense of the stability of the native state (4). The extent to which protein folding rates are under selective pressure can potentially be resolved by comparing the folding rates of sequences not obtained through natural selection to the folding rates of naturally occurring proteins; if kinetic accessibility is under selective pressure, the folding rates of the laboratory-generated sequences should not be restricted to the time scales typically observed for small proteins.
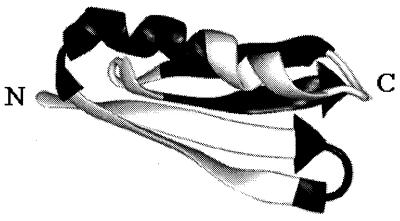
Herein we use such an approach to determine the extent to which the folding rate of the IgG binding domain of peptostreptococcal protein L has been optimized by natural selection. The kinetics and thermodynamics of folding of a tryptophan-containing mutant of protein L (hereafter referred to as wild type) have been extensively characterized (5, 6), and a phage-display selection method (7) has been developed that allows the retrieval of functional folded variants from combinatorial libraries. The high-resolution NMR solution structure of protein L consists of a helix packed against a four-stranded β-sheet with the order of the secondary structure elements ββαββ (Fig. 1) (8).
Figure 1.
Protein L structure. Starting from the N terminus, alternating white and black segments correspond to residues randomized in strand 1 (Val-4 to Phe-12, white) and turn 1 (Phe-12 to Ser-16, black), binding residues in strand 2 (white), residues randomized in the helix (Gly-24 to Tyr-34, black), binding residues in the helix (white), residues not subjected to mutagenesis in strand 3 (black), and residues randomized in turn 2 (Ala-52 to Thr-57, white) and strand 4 (Leu-58 to Ala-63, black). The image was created with midasplus.
We describe the folding thermodynamics and kinetics of functional folded variants retrieved from combinatorial libraries of highly mutated protein L sequences by using the phage-display selection method. Although all of the variants had reduced stabilities, half had folding rates faster than wild type. This observation suggests that despite the enormous search space, the rapid folding of biological proteins is not the result of direct natural selection; instead, fast folding appears to be an intrinsic property of polypeptide chains that adopt stable native states.
MATERIALS AND METHODS
All reagents, solutions, and enzymes for molecular biology procedures were as described (7). Phagemid libraries were constructed in which the two β-hairpin turns (9), helix, and the two inner strands (1 and 4) were independently replaced by randomized synthetic cassettes (Fig. 1). Libraries for the strands and the helix were constructed as described for the turn libraries (9) using oligonucleotides with sequences 5′-GCTCAGGCGGCCATGGAADHMDHMDHMDHMDHMDHMDHMDHMDHMGCAAATGGGTC-3′ and 5′-GTTCCTTTGAATTCCGCAGTTTGTGTGGACCCATTTCG-3′ for strand 1, 5′-GATAAAGGTTATACTDHWDHWDHWDHWDHWDHWGGATAGATGCAC-3′ and 5′-ACGCGTTTCCTCCGTGCATCTATCC-3′ for strand 4, and 5′-GGGGGGGGAATTCAAANNSNNSNNSNNSNNSNNSNNSNNSNNSGCATATGCAGACGC-3′ and 5′-GCGTCTGCATAT-3′ for the helix (where D = A, G or T; H = A, C or T; M = A or C; N = A, C, G, or T; S = G or C; and W = A or T). An additional PCR-amplification step was used for strand 4 and the helix using primers with sequences 5′-GGAGAATGGACTGTCGACGTTGCAGATAAAGGTTATACT-3′ and 5′-GTCACCCTCAGCACACGCGTTTCCTCC-3′ for strand 4, and 5′-GGGGGGGGAATTC-3′ and the shorter oligonucleotide described above for the helix. The resulting double-stranded DNA products were digested with NcoI and EcoRI for strand 1, SalI and MluI for strand 4, and EcoRI and NdeI for the helix. The phage-display selection and colony-lift screening were carried out as described (7). Standard M13 dye primer cycle sequencing protocols (Perkin–Elmer) were used for DNA sequencing. The expression, purification, and thermodynamic and kinetic characterization of the folded variants retrieved from the libraries were carried out as described (6, 9). The sequence and kinetic data for the variants retrieved from the turn 1 and turn 2 libraries were reported in ref. 9.
RESULTS
The principle behind the phage-display selection is that the binding of protein L to IgG requires that the protein be properly folded; this is expected because the IgG binding residues are not contiguous in sequence. We previously showed that the phage display selection indeed yields properly folded variants of protein L: variants with widely divergent sequences retrieved by using the selection had native-like CD and one-dimensional NMR spectra, whereas a single point mutant that disrupted folding was strongly selected against (7). Before setting out to characterize the range of folding times in heavily mutated sequences retrieved in the phage selection, it was first critical to determine whether the phage selection had any kinetic bias.
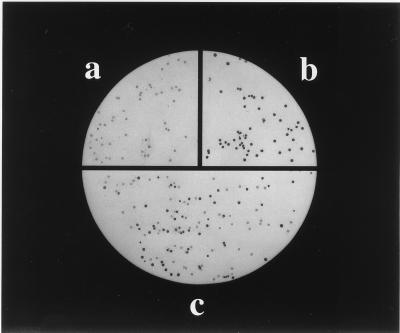
To investigate the possibility of such bias, we took advantage of glycine to alanine point mutations in the two turns that have similar stabilities but different folding rates (9). Biopanning of homogeneous phagemid populations suggested that the selection had relatively little bias: the recovery of phagemids displaying the G15A mutant, which folds nearly 10-fold more slowly than wild type, was similar to the recovery of phage displaying the equally stable but more rapidly folding G55A mutant and phage displaying wild-type protein L (Table 1). The possibility of bias was further probed by using competition experiments in which an equal number of wild-type and G15A phagemids were combined and then subjected to biopanning. Escherichia coli colonies harboring the G15A phagemid gave a somewhat weaker signal in the colony-lift screening for IgG binding than did colonies harboring the wild-type phagemid [Fig. 2, compare G15A (a) and wild-type (b)], and thus the recovery of the two types of phagemids in the competition experiment could be assessed with the colony lift assay (Fig. 2c). Importantly, the recoveries of the two types of phagemids were very similar (Table 1 and Fig. 2c), showing that the selection procedure does not distinguish between rapidly and slowly folding variants.
Table 1.
Biopanning does not distinguish between variants with different folding rates
| Bluescript | Wild type | G15A | G55A | Wild-type G15A mixture
|
Wild-type G55A mixture
|
|||
|---|---|---|---|---|---|---|---|---|
| Wild type | G15A | Wild type | G55A | |||||
| In | 108 | 108 | 108 | 108 | 108 | 108 | 108 | 108 |
| Out | 60 | 3.6 × 105 | 5.4 × 105 | 2.6 × 105 | 1.3 × 105 | 1.4 × 105 | 1.4 × 105 | 1.1 × 105 |
| Recovery, % | 0.00006 | 0.36 | 0.54 | 0.26 | 0.13 | 0.14 | 0.14 | 0.11 |
Individual phagemids (Bluescript, wild type, G15A, and G55A) or mixtures of wild-type and G15A or G55A mutant phagemids were subjected to biopanning as described (5). In, number of phagemids added to IgG coated magnetic beads; out, number eluted after extensive washes. The folding rates for wild type, G15A, and G55A are 61, 7, and 36 s−1, respectively. The Bluescript control phagemids do not display protein L.
Figure 2.
Phage selection is not biased toward more rapidly folding sequences. Colony-lift assay of XL1-Blue colonies infected with G15A phagemids (a), wild-type protein L phagemids (b), and phagemids eluted from IgG coated beads incubated with equal amounts of wild type and G15A phagemids (c) (Table 1). Although the refolding rate of G15A is nearly 10-fold slower than wild type, the recoveries of the two phagemids are similar in the competition biopanning assay.
To obtain protein L variants with large numbers of sequence changes in different parts of the protein, we selected functional folded variants of protein L from phagemid libraries in which the two β-hairpin turns (9), the helix, and strands 1 and 4 were independently randomized (Fig. 1). Thirty-six of the 62 residues in protein L were subjected to mutagenesis; residues involved in IgG binding (10) along with the third strand were not randomized. The sequences of variants that exhibited IgG binding activity in the biopanning selection and colony-lift screening were highly divergent (Table 2). All 36 positions subjected to mutagenes tolerated a non-wild-type residue, and at least 50% of the residues randomized in each variant were changed. Strand 1, the helix, and the two turns tolerated complete mutagenesis, whereas strand 2 tolerated simultaneous changes of 5 of the 6 residues.
Table 2.
Sequence changes in the functional fariants obtained from the phage selection and screen
| Strand 1
|
Turn 1
|
Helix
|
Turn 2
|
Strand 4
|
|||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vt | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 12 | 13 | 14 | 15 | 16 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | ||||||||
| V | T | I | K | A | N | L | I | F | Vt | F | A | N | G | S | Vt | G | T | F | E | K | A | T | S | E | A | Y | Vt | A | D | K | G | Y | T | Vt | L | N | I | K | F | A | |||||
| b1-a | S | V | V | T | T | Y | Y | F | L | t1-a | D | C | Q | h-a | L | G | W | E | L | V | N | L | W | t2-a | P | T | S | A | T | Q | b4-a | I | Y | F | T | Y | |||||||||
| b1-b | D | V | S | V | I | Y | V | L | t1-b | P | C | A | h-b | Q | L | L | F | L | M | D | L | V | t2-b | L | T | P | T | Q | b4-b | Y | T | T | |||||||||||||
| b1-c | E | L | V | Y | Y | T | Y | Q | P | C | Q | P | h-c | E | Y | H | T | L | Y | Q | I | W | t2-c | Q | A | Q | R | I | T | Y | V | Y | T | ||||||||||||
| E | I | V | V | V | D | Y | V | L | T | P | C | Q | N | h-d | S | G | L | E | L | G | S | V | L | N | Q | S | A | R | L | F | F | S | Y | L | |||||||||||
| E | V | V | V | Y | Y | S | L | L | P | C | S | R | T | D | L | N | R | C | L | L | D | V | I | L | P | A | E | S | L | S | F | A | Y | S | |||||||||||
| E | S | V | V | Y | Y | E | L | L | P | C | Q | G | A | Q | G | T | P | P | Y | L | D | V | W | S | P | S | S | E | L | V | T | F | Y | E | |||||||||||
| D | V | V | I | V | T | L | S | P | C | M | P | P | T | D | L | V | W | A | Q | L | W | T | P | T | P | Q | E | I | Y | F | I | N | |||||||||||||
| T | S | V | T | V | Y | Y | G | P | C | L | P | Q | L | P | S | V | V | E | Q | M | I | Q | R | P | H | G | V | V | S | F | Y | L | |||||||||||||
| S | V | S | Y | I | T | L | P | C | S | D | G | P | V | E | V | V | W | D | L | V | T | P | T | P | Q | E | V | F | N | Y | I | ||||||||||||||
| D | V | Y | I | T | T | T | P | C | Q | G | I | G | W | Q | V | R | Q | S | V | V | D | K | R | S | Q | N | S | F | N | Y | V | ||||||||||||||
| E | S | V | V | K | S | L | P | C | Q | E | G | L | Q | A | S | L | T | Q | L | W | S | H | D | L | H | A | I | F | F | N | N | ||||||||||||||
| T | V | T | Y | Y | N | T | P | C | L | P | K | R | T | E | V | F | R | Q | V | C | Q | E | T | L | G | L | F | F | N | Y | T | ||||||||||||||
| E | V | I | F | E | L | L | P | C | H | Q | P | I | F | V | V | L | E | Q | V | I | H | Q | D | M | R | E | V | F | T | Y | T | ||||||||||||||
| E | V | Y | Y | L | L | V | P | C | R | E | R | G | S | E | V | F | G | L | V | F | G | G | G | Q | E | S | F | F | S | Y | |||||||||||||||
| D | I | V | Y | V | F | L | P | S | E | Q | G | A | P | Y | D | S | I | W | L | P | R | E | H | M | F | F | N | I | |||||||||||||||||
| E | V | Y | V | Y | L | V | P | C | Q | M | P | L | Q | F | A | Q | L | L | G | P | G | R | Q | F | V | Y | I | ||||||||||||||||||
| T | S | T | T | I | L | P | C | Q | P | A | V | V | V | G | R | V | L | M | P | S | L | E | Y | Y | Y | N | |||||||||||||||||||
| S | E | V | F | V | I | P | S | P | N | L | V | V | L | Q | Q | V | V | M | A | Q | S | E | Y | I | Y | K | |||||||||||||||||||
| S | F | T | F | T | Y | P | C | P | S | L | K | N | L | V | P | I | L | Q | P | Q | D | R | V | F | S | L | |||||||||||||||||||
| E | Y | V | T | Y | Y | P | C | L | G | G | L | G | W | E | Q | V | W | L | P | N | Q | G | T | Y | Y | I | |||||||||||||||||||
| V | T | Y | Y | V | P | E | A | D | R | Q | G | V | Y | A | Q | V | F | Q | P | T | A | I | V | I | Y | ||||||||||||||||||||
| T | V | T | I | Y | L | P | G | C | P | L | G | S | S | L | Q | L | G | P | Q | P | Y | Y | V | ||||||||||||||||||||||
| A | V | Y | Y | Y | P | C | Q | S | P | I | L | R | L | V | S | Q | F | M | T | V | T | ||||||||||||||||||||||||
| P | E | S | Q | G | S | P | V | Y | T | ||||||||||||||||||||||||||||||||||||
| P | E | C | V | S | Q | Q | |||||||||||||||||||||||||||||||||||||||
Wild-type sequence and residue numbers are indicated at the top. The variants (Vt) selected for biophysical characterization are labeled to the left of the sequences. t2-b has a single residue deletion.
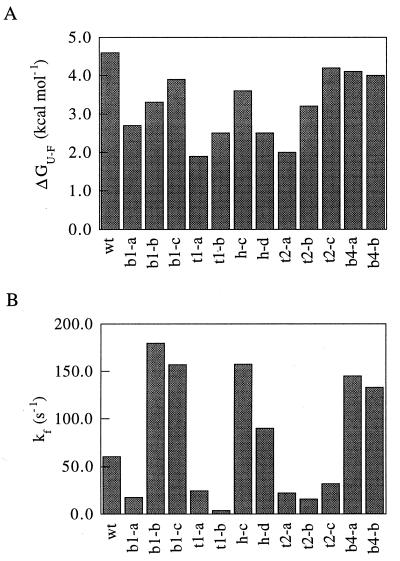
To determine the range of kinetic and thermodynamic properties in the mutant population, 14 variants with substantial sequence changes were subcloned, expressed, and purified for biophysical characterization (Table 2). Far-UV CD spectra of most of the mutants were similar to wild type with the exception of those of helix mutants h-a, h-c, and h-d, which displayed a significant decrease in negative ellipticity near 220 nm that was likely due to a partial loss of helical structure in the randomized region (data not shown). With the exception of h-a and h-b, the variants displayed cooperative denaturation transitions in guanidine hydrochloride (GuHCl) denaturation experiments, which were fit well by a two-state model of folding (data not shown). The stabilities of the variants, which ranged from 1.9 to 4.2 kcal/mol (1 cal = 4.184 J), were all less than wild type (4.6 kcal/mol; Fig. 3A).
Figure 3.
Distributions of free energies of unfolding and refolding rates in the randomized variants. (A) Free energies of unfolding. Free energies of unfolding in H2O were obtained by determining the folding equilibrium constant through fluorescence measurements at different concentrations of GuHCl and then extrapolating to zero molar denaturant (6). (B) Refolding rates in the absence of denaturant. Folding rates in a broad range of GuHCl concentrations were determined by rapidly mixing denatured protein in 3 M GuHCl with buffer containing various concentrations of GuHCl. The folding rates in water were estimated by linear extrapolation from the logarithms of the folding rates in 0.3–2.0 M GuHCl.
The folding and unfolding rates of the 14 variants were determined by using stopped-flow fluorescence over a broad range of GuHCl concentrations. The kinetic data for 12 of the variants fit well to a two-state model of folding (data not shown). The folding rates in the mutant population ranged from 4 to 180 s−1, compared with 61 s−1 for wild type (Fig. 3B). Although mutants b1-a, h-c, and h-d had nine residue changes, their folding rates were within 4-fold of wild type. Interestingly, 50% of the variants had a folding rate greater than wild type. Only one mutant, t1-b, had a folding rate as slow as the G15A point mutant.
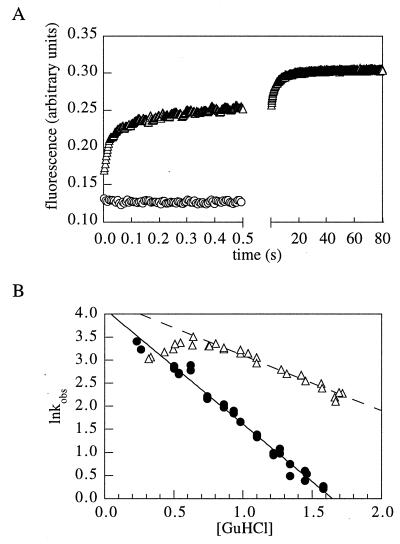
More complex kinetics were observed for two of the variants with many changes in the helix. Variant h-b displayed multiexponential refolding kinetics (Fig. 4A), and the logarithm of the folding rate of h-a vs. GuHCl concentration was not linear at low GuHCl concentrations (Fig. 4B). The complex kinetics of h-b, the very slow phase in particular, is likely to be at least in part due to dimerization because the protein was found to be a dimer by gel filtration chromatography in the absence of denaturant (data not shown). The decrease in folding rate of h-a at low GuHCl concentrations suggests the formation of kinetically trapped states under these conditions (Fig. 4B). A similar result was found for human spliceosomal protein U1A and was attributed to transient aggregation of the denatured protein under low denaturant refolding conditions (11).
Figure 4.
Anomalous refolding kinetics of variants h-a and h-b. (A) Kinetics of refolding of h-b (triangles, 0.4 M, final GuHCl concentration). The signal of the unfolded protein in 4 M GuHCl is indicated by circles. (B) GuHCl dependence of the logarithm of the folding rate constant for h-a (▵) and wild type (•). The deviation from linearity below 0.7 M GuHCl suggests the accumulation of an intermediate.
DISCUSSION
Our results suggest that the sequences of naturally occurring proteins have not been extensively optimized for rapid folding by natural selection. Large sequence changes, although almost invariably reducing the stability of protein L (Fig. 3A), increased the folding rate as often as decreasing it (Fig. 3B). Importantly, the phage-display selection procedure that was used to retrieve the variants had no detectable bias against slowly folding sequences (Table 1 and Fig. 2).
Because Keq = kf/ku for simple two-state folding reactions, there is a limit to how slow the folding rate can be without completely destabilizing the protein in the absence of simultaneous decreases in the unfolding rate. However, the results with the G15A mutant show that the folding rate can be decreased substantially without disrupting the efficiency of phage recovery in the selection. The fact that very few of the random variants had folding rates as slow as G15A suggests that there are not strong kinetic constraints throughout the majority of the sequence. The most dramatic departure from the wild-type folding kinetics was observed for two variants that were likely to partially aggregate during folding (Fig. 4); while not apparently optimizing the folding rate, natural selection does appear to select against aggregation during folding because such behavior is rare for small protein domains.
The idea that folding rates are not directly optimized by natural selection is also suggested by our studies of the folding kinetics of heavily simplified variants of the src homology 3 (SH3) domain obtained with a phage-display selection similar to that used herein (12). Highly mutated variants of the SH3 domain in which 40 of the 57 residues were simplified to isoleucine, lysine, glutamic acid, alanine, or glycine had folding rates almost identical to wild type. Thus, the protein L and SH3 results strongly suggest that the fast folding rates typical of small biological proteins are not directly optimized by natural selection.
As mentioned in the Introduction, there are a number of folding reactions with large kinetic barriers. For example, the serine proteases subtilisin and α-lytic protease are synthesized with large pro regions that are required for proper folding: in the absence of the pro region, folding does not occur in vitro or in vivo (2, 3). There are two possible explanations for this inability to fold: either the removal of the pro region exposes ubiquitous free-energy barriers on polypeptide chain folding free-energy landscapes that are avoided in naturally occurring proteins due to natural selection or the large free-energy barriers are an evolutionary specialization specific to the proteases. Our results with protein L and the SH3 domain are consistent with the second explanation because large free-energy barriers are not encountered when the sequences are substantially perturbed from those of the naturally occurring proteins. The evolutionary specialization hypothesis is also consistent with the fact that the proteases are involved in amino acid scavenging in harsh extracellular environments, and hence in contrast to most intracellular proteins, there is likely to be strong selective pressure to maximize their lifetimes. Selection for a large barrier to unfolding would accomplish this at the cost of a large free-energy barrier to folding; the pro regions could serve to transiently reduce the barriers during folding (both pro regions are extremely protease sensitive and are probably destroyed shortly after folding is complete).
Why might kinetic accessibility be a consequence of thermodynamic stability for small protein domains? A likely answer is that the interactions that stabilize the native state also stabilize conformations with partially formed native structure, and thus the energy on average decreases with increasing native structure. The result is the funnel-shaped energy landscape figured prominently in recent theoretical treatments of folding (13–15). An explanation for our results is that the phage selection selects for sequences stable in the native state and consequently for a funnel-shaped energy landscape. An important implication of the results presented herein is that protein design efforts can focus on optimizing the stability of a particular folded conformation without explicitly designing for kinetic accessibility; this is encouraging for protein design because the sequence determinants of protein stability are much better understood than those for folding kinetics.
There are interesting parallels between our results and those obtained in theoretical studies of folding. In a study of 27-residue chains on the simple cubic lattice, sequences were optimized in the context of a simple energy function for stability in the native state (16). As in our experiments, kinetic accessibility was found to be a consequence of selection for stability: a large fraction of the optimized sequences, but not unselected sequences, folded readily to their native states in kinetics simulations. In the analytical theory of folding developed in ref. 17, folding occurs rapidly when the folding transition temperature Tf is higher than the glass transition temperature Tg. The simple two-state behavior of the folding reactions of most small naturally occurring proteins and the temperature dependence of these reactions suggest that Tg is far below the range accessible to experiment for naturally occurring proteins; our results suggest that this is a consequence of polypeptide-chain physical chemistry rather than evolutionary selection. The roughness of the free-energy landscape and, hence, Tg are likely to depend on basic properties not significantly altered in our experiments such as the amino acid composition and the dispersion in the interresidue interaction energies.
Although the combined random sequence selection and kinetic analysis approach taken in this article was specifically designed to address the issue of evolutionary optimization of folding rates, the results also provide valuable clues about the mechanism of protein L folding. The dramatic difference in sequence conservation in the two turns and the 10-fold slow down in the folding rate of mutant t1-b led to targeted mutagenesis experiments that showed that the first turn but not the second turn is formed at the rate-limiting step in folding (9). A second striking feature of the results in Table 2 and Fig. 3B is the correlation between increases in the size of hydrophobic core residues and increases in folding rate. The inference that the rate limiting step in folding also involves hydrophobic core assembly is currently being tested.
Acknowledgments
We thank David Shortle, Peter Wolynes, and members of the Baker group for helpful comments on the manuscript and Keith Zachrone for assistance with DNA sequencing. This work was supported by a National Institutes of Health grant (GM5188) and young investigator awards to D.B. from the National Science Foundation and the Packard Foundation.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: GuHCl, guanidine hydrochloride; SH3, src homology 3.
References
- 1.Levinthal C. J Chim Phys. 1968;65:44–45. [Google Scholar]
- 2.Baker D, Agard D A. Biochemistry. 1994;33:7505–7509. doi: 10.1021/bi00190a002. [DOI] [PubMed] [Google Scholar]
- 3.Strausberg S, Alexander P, Wang L, Schwarz F, Bryan P. Biochemistry. 1993;32:8112–8119. doi: 10.1021/bi00083a009. [DOI] [PubMed] [Google Scholar]
- 4.Ladurner A G, Itzhaki L S, Fersht A R. Fold Des. 1997;2:363–368. doi: 10.1016/S1359-0278(97)00050-3. [DOI] [PubMed] [Google Scholar]
- 5.Yi Q, Baker D. Protein Sci. 1996;5:1060–1066. doi: 10.1002/pro.5560050608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scalley M L, Yi Q, Gu H, McCormack A, Yates J R, Baker D. Biochemistry. 1997;36:3373–3382. doi: 10.1021/bi9625758. [DOI] [PubMed] [Google Scholar]
- 7.Gu H, Yi Q, Bray S T, Riddle D S, Shiau A K, Baker D. Protein Sci. 1995;4:1108–1117. doi: 10.1002/pro.5560040609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wikstrom, M., Sjobring, U., Kastern, W., Bjorck, L., Drakenberg, T. & Forsen, S. Biochemistry 32, 3381–3386. [DOI] [PubMed]
- 9.Gu H, Kim D, Baker D. J Mol Biol. 1997;274:588–596. doi: 10.1006/jmbi.1997.1374. [DOI] [PubMed] [Google Scholar]
- 10.Wikstrom M, Sjobring U, Drakenberg T, Forsen S, Bjorck L. J Mol Biol. 1995;250:128–133. doi: 10.1006/jmbi.1995.0364. [DOI] [PubMed] [Google Scholar]
- 11.Silow M, Oliveberg M. Proc Natl Acad Sci USA. 1997;94:6084–6086. doi: 10.1073/pnas.94.12.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riddle D S, Santiago J V, Bray-Hall S T, Doshi N, Grantcharova V P, Yi Q, Baker D. Nat Struct Biol. 1997;4:805–809. doi: 10.1038/nsb1097-805. [DOI] [PubMed] [Google Scholar]
- 13.Leopold P E, Montal M, Onuchic J N. Proc Natl Acad Sci USA. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryngelson J D, Onuchic J N, Socci N D, Wolynes P G. Proteins Struct Funct Genet. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 15.Dill K A, Chan H S. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 16.Shakhnovich E I, Gutin A M. Proc Natl Acad Sci USA. 1993;90:7195–7199. doi: 10.1073/pnas.90.15.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryngelson J D, Wolynes P G. J Phys Chem. 1989;93:6902–6915. [Google Scholar]