Abstract
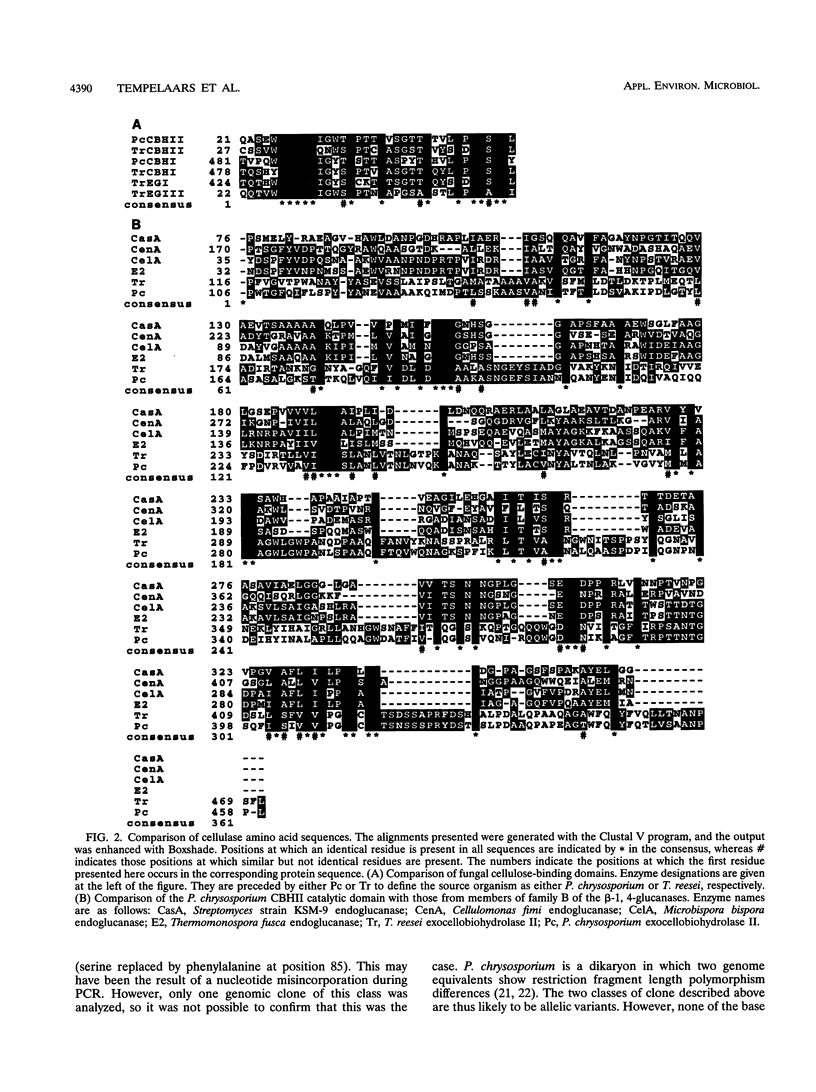
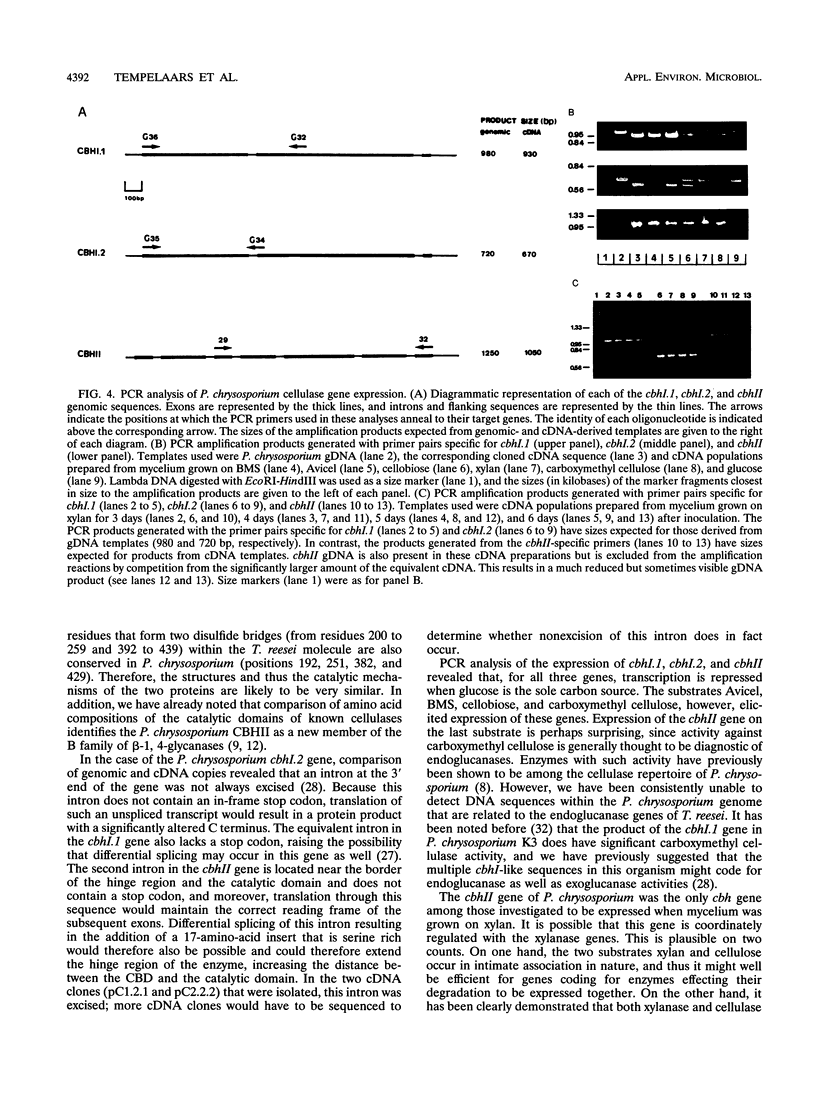
Two cDNA sequences representing putative allelic variants of the Phanerochaete chrysosporium cbhII gene were isolated by hybridization to the Trichoderma reesei cbhII gene. Both of the equivalent genomic sequences were subsequently isolated by the inverse PCR technique. DNA sequencing showed that the cbhII open reading frame of 1,380 bp codes for a putative polypeptide of 460 amino acids which is interrupted by six introns. The domain structure found in T. reesei cbhII is conserved in the equivalent P. chrysosporium protein. The overall similarity between the two gene products is 54%, with the region of highest conservation being found in the cellulose-binding domain (65%). Unlike the cbhI gene of P. chrysosporium, cbhII does not appear to be a member of a class of closely related genes. CBHII is a new member of family B of the beta-1, 4-glucanases. Alignment of the P. chrysosporium and T. reesei CBHII protein sequences showed that all of the residues important for the formation of the extended loops of the catalytic domain and those residues that are involved in the catalytic action of the T. reesei enzyme are also present in the P. chrysosporium equivalent. The profiles of cbh gene expression in P. chrysosporium reveal that while cbhI.1 and cbhI.2 could be coregulated, cbhII can be independently controlled. The latter is so far the only cellulase gene found to be expressed when the fungus is grown on oat spelt arabinoxylan, suggesting that it may play an active role in the xylanolytic as well as the cellulolytic systems.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brooks P., Sims P., Broda P. Isozyme specific polymerase chain reaction analysis of differential gene expression: a general method applied to lignin peroxidase genes of Phanerochaete chrysosporium. Biotechnology (N Y) 1993 Jul;11(7):830–834. doi: 10.1038/nbt0793-830. [DOI] [PubMed] [Google Scholar]
- Brown A., Sims P. F., Raeder U., Broda P. Multiple ligninase-related genes from Phanerochaete chrysosporium. Gene. 1988 Dec 15;73(1):77–85. doi: 10.1016/0378-1119(88)90314-9. [DOI] [PubMed] [Google Scholar]
- Covert S. F., Bolduc J., Cullen D. Genomic organization of a cellulase gene family in Phanerochaete chrysosporium. Curr Genet. 1992 Nov;22(5):407–413. doi: 10.1007/BF00352442. [DOI] [PubMed] [Google Scholar]
- Covert S. F., Vanden Wymelenberg A., Cullen D. Structure, organization, and transcription of a cellobiohydrolase gene cluster from Phanerochaete chrysosporium. Appl Environ Microbiol. 1992 Jul;58(7):2168–2175. doi: 10.1128/aem.58.7.2168-2175.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert K. A., Kunkel T. A. High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1990 Jul 11;18(13):3739–3744. doi: 10.1093/nar/18.13.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson K. E., Pettersson B. Extracellular enzyme system utilized by the fungus Sporotrichum pulverulentum (Chrysosporium lignorum) for the breakdown of cellulose. 1. Separation, purification and physico-chemical characterization of five endo-1,4-beta-glucanases. Eur J Biochem. 1975 Feb 3;51(1):193–206. doi: 10.1111/j.1432-1033.1975.tb03919.x. [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey B. J., Mayfield M. B., Brown J. A., Gold M. H. Characterization of a gene encoding a manganese peroxidase from Phanerochaete chrysosporium. Gene. 1990 Sep 1;93(1):119–124. doi: 10.1016/0378-1119(90)90144-g. [DOI] [PubMed] [Google Scholar]
- Henrissat B., Claeyssens M., Tomme P., Lemesle L., Mornon J. P. Cellulase families revealed by hydrophobic cluster analysis. Gene. 1989 Sep 1;81(1):83–95. doi: 10.1016/0378-1119(89)90339-9. [DOI] [PubMed] [Google Scholar]
- James C. M., Felipe M. S., Sims P. F., Broda P. Expression of a single lignin peroxidase-encoding gene in Phanerochaete chrysosporium strain ME446. Gene. 1992 May 15;114(2):217–222. doi: 10.1016/0378-1119(92)90577-c. [DOI] [PubMed] [Google Scholar]
- Kirk T. K., Farrell R. L. Enzymatic "combustion": the microbial degradation of lignin. Annu Rev Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- Penttilä M., Lehtovaara P., Nevalainen H., Bhikhabhai R., Knowles J. Homology between cellulase genes of Trichoderma reesei: complete nucleotide sequence of the endoglucanase I gene. Gene. 1986;45(3):253–263. doi: 10.1016/0378-1119(86)90023-5. [DOI] [PubMed] [Google Scholar]
- Pribnow D., Mayfield M. B., Nipper V. J., Brown J. A., Gold M. H. Characterization of a cDNA encoding a manganese peroxidase, from the lignin-degrading basidiomycete Phanerochaete chrysosporium. J Biol Chem. 1989 Mar 25;264(9):5036–5040. [PubMed] [Google Scholar]
- Raeder U., Broda P. Meiotic segregation analysis of restriction site polymorphisms allows rapid genetic mapping. EMBO J. 1986 Jun;5(6):1125–1127. doi: 10.1002/j.1460-2075.1986.tb04336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeder U., Thompson W., Broda P. RFLP-based genetic map of Phanerochaete chrysosporium ME446: lignin peroxidase genes occur in clusters. Mol Microbiol. 1989 Jul;3(7):911–918. doi: 10.1111/j.1365-2958.1989.tb00240.x. [DOI] [PubMed] [Google Scholar]
- Rouvinen J., Bergfors T., Teeri T., Knowles J. K., Jones T. A. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science. 1990 Jul 27;249(4967):380–386. doi: 10.1126/science.2377893. [DOI] [PubMed] [Google Scholar]
- Saloheimo M., Lehtovaara P., Penttilä M., Teeri T. T., Ståhlberg J., Johansson G., Pettersson G., Claeyssens M., Tomme P., Knowles J. K. EGIII, a new endoglucanase from Trichoderma reesei: the characterization of both gene and enzyme. Gene. 1988;63(1):11–22. doi: 10.1016/0378-1119(88)90541-0. [DOI] [PubMed] [Google Scholar]
- Sims P. F., Soares-Felipe M. S., Wang Q., Gent M. E., Tempelaars C., Broda P. Differential expression of multiple exo-cellobiohydrolase I-like genes in the lignin-degrading fungus Phanerochaete chrysosporium. Mol Microbiol. 1994 Apr;12(2):209–216. doi: 10.1111/j.1365-2958.1994.tb01010.x. [DOI] [PubMed] [Google Scholar]
- Sims P., James C., Broda P. The identification, molecular cloning and characterisation of a gene from Phanerochaete chrysosporium that shows strong homology to the exo-cellobiohydrolase I gene from Trichoderma reesei. Gene. 1988 Dec 30;74(2):411–422. doi: 10.1016/0378-1119(88)90174-6. [DOI] [PubMed] [Google Scholar]
- Teeri T. T., Lehtovaara P., Kauppinen S., Salovuori I., Knowles J. Homologous domains in Trichoderma reesei cellulolytic enzymes: gene sequence and expression of cellobiohydrolase II. Gene. 1987;51(1):43–52. doi: 10.1016/0378-1119(87)90472-0. [DOI] [PubMed] [Google Scholar]
- Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., Claeyssens M. Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur J Biochem. 1988 Jan 4;170(3):575–581. doi: 10.1111/j.1432-1033.1988.tb13736.x. [DOI] [PubMed] [Google Scholar]
- Uzcategui E., Ruiz A., Montesino R., Johansson G., Pettersson G. The 1,4-beta-D-glucan cellobiohydrolases from Phanerochaete chrysosporium. I. A system of synergistically acting enzymes homologous to Trichoderma reesei. J Biotechnol. 1991 Jul;19(2-3):271–285. doi: 10.1016/0168-1656(91)90064-3. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]