Abstract
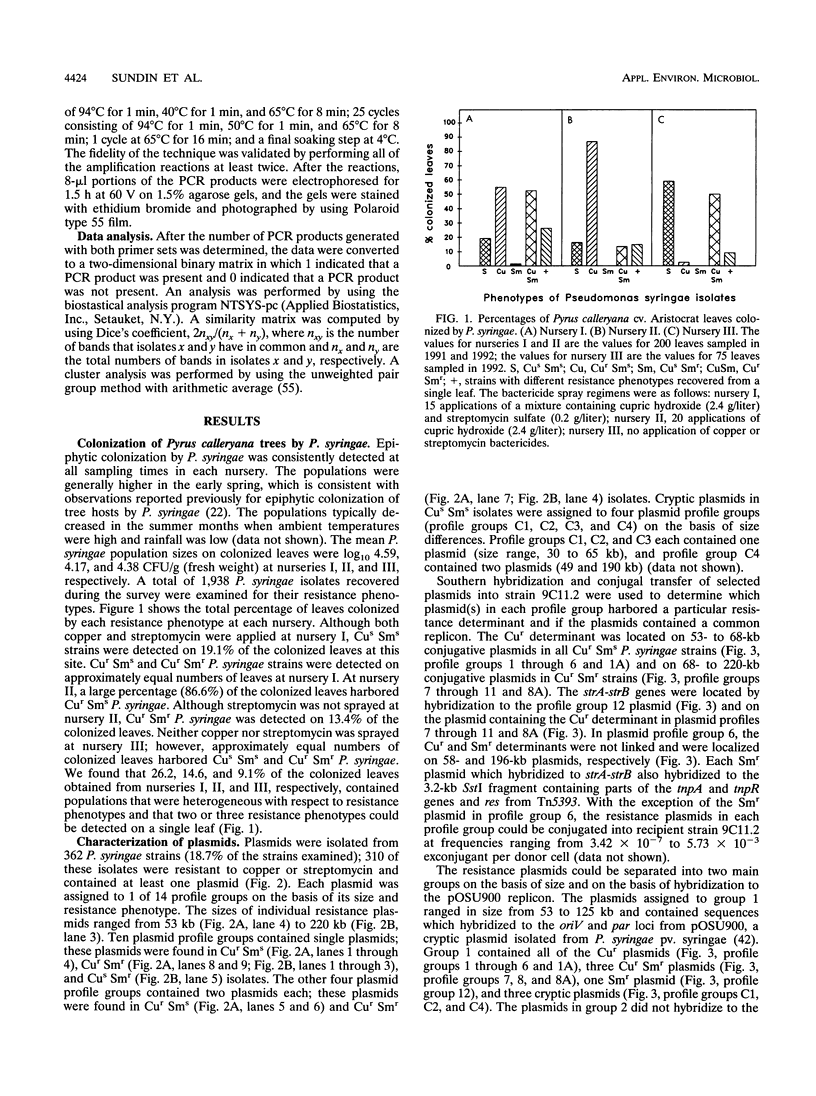
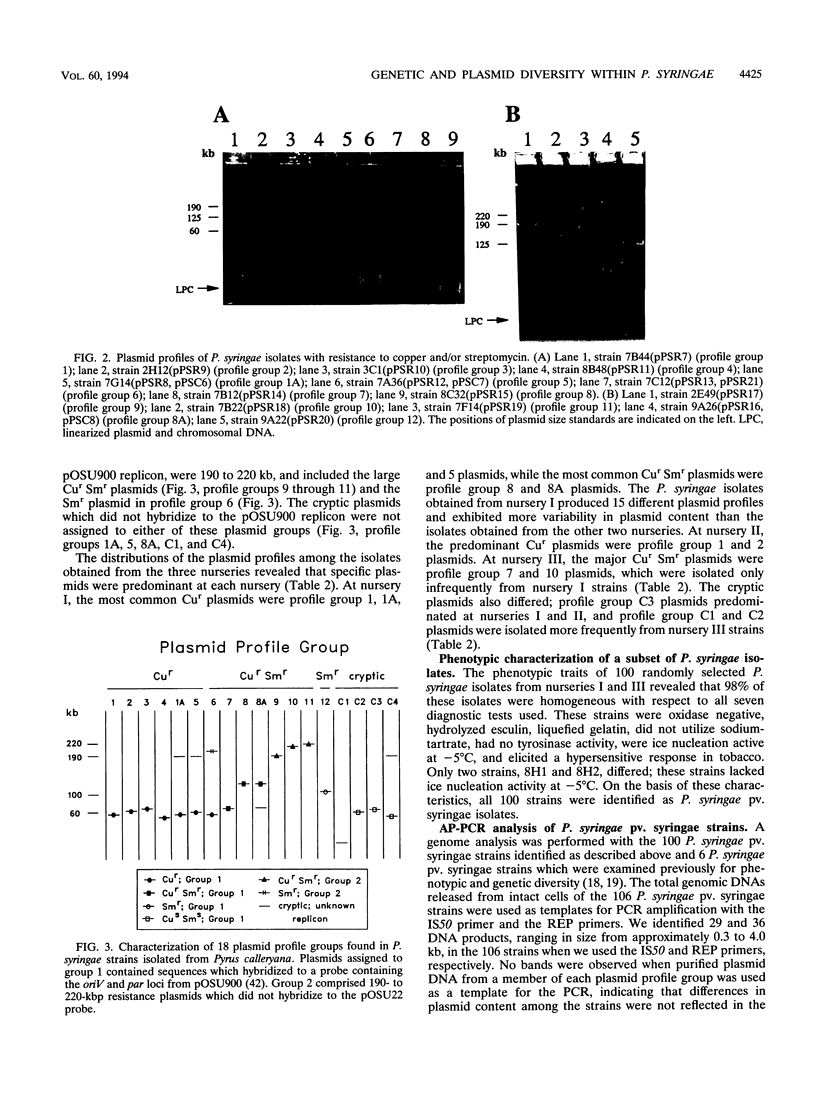
We examined the genetic and plasmid diversity within natural populations of Pseudomonas syringae isolated from three ornamental pear nurseries in eastern Oklahoma. The bactericide spray regimen differed at each nursery; copper and streptomycin, only copper, and no bactericides were applied at nurseries I, II, and III respectively. Resistance to copper (Cur) and resistance to streptomycin (Smr) were determined for 1,938 isolates of P. syringae; isolates from nurseries I and II were generally Cur Sms; whereas most isolates from nursery III were Cus Sms. The plasmid profiles of 362 isolates were determined, and six, one, seven, and four plasmid profiles were obtained for Cur, Smr, Cur Smr, and Cus Sms isolates, respectively. All Smr plasmids contained sequences homologous to the strA and strB Smr genes from broad-host-range plasmid RSF1010 and were associated with Smr transposon Tn5393. Plasmids were placed into two groups on the basis of hybridization to the oriV and par sequences from pOSU900, a cryptic plasmid in P. syringae pv. syringae. A total of 100 randomly chosen P. syringae isolates from nurseries I and III were analyzed for genetic diversity by using the arbitrarily primed PCR (AP-PCR) technique. An analysis of chromosomal genotypes by AP-PCR revealed a high degree of genetic diversity among the isolates, and the results of this analysis indicated that the isolates could be clustered into two distinct groups. The plasmid profiles were specific to isolates belonging to particular AP-PCR groups. Within each AP-PCR group, identical plasmid profiles were produced by isolates that had different chromosomal genotypes, implying that plasmid transfer has played an important role in the dissemination of Cur and Smr within the populations studied.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender C. L., Cooksey D. A. Indigenous plasmids in Pseudomonas syringae pv. tomato: conjugative transfer and role in copper resistance. J Bacteriol. 1986 Feb;165(2):534–541. doi: 10.1128/jb.165.2.534-541.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender C. L., Cooksey D. A. Molecular cloning of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1987 Feb;169(2):470–474. doi: 10.1128/jb.169.2.470-474.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzar H., Ouadah D., Krimi Z., Jones J. B., Trovato M., Petit A., Dessaux Y. Correlative Association between Resident Plasmids and the Host Chromosome in a Diverse Agrobacterium Soil Population. Appl Environ Microbiol. 1993 May;59(5):1310–1317. doi: 10.1128/aem.59.5.1310-1317.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou C. S., Jones A. L. Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other gram-negative bacteria. J Bacteriol. 1993 Feb;175(3):732–740. doi: 10.1128/jb.175.3.732-740.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D. A., Azad H. R., Cha J. S., Lim C. K. Copper resistance gene homologs in pathogenic and saprophytic bacterial species from tomato. Appl Environ Microbiol. 1990 Feb;56(2):431–435. doi: 10.1128/aem.56.2.431-435.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D. A. Characterization of a Copper Resistance Plasmid Conserved in Copper-Resistant Strains of Pseudomonas syringae pv. tomato. Appl Environ Microbiol. 1987 Feb;53(2):454–456. doi: 10.1128/aem.53.2.454-456.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D. A. Copper uptake and resistance in bacteria. Mol Microbiol. 1993 Jan;7(1):1–5. doi: 10.1111/j.1365-2958.1993.tb01091.x. [DOI] [PubMed] [Google Scholar]
- Cooksey D. A. Plasmid-Determined Copper Resistance in Pseudomonas syringae from Impatiens. Appl Environ Microbiol. 1990 Jan;56(1):13–16. doi: 10.1128/aem.56.1.13-16.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercolani G. L. Variability among isolates of Pseudomonas syringae pv. savastanoi from the phylloplane of the olive. J Gen Microbiol. 1983 Apr;129(4):901–916. doi: 10.1099/00221287-129-4-901. [DOI] [PubMed] [Google Scholar]
- Garde S., Bender C. L. DNA probes for detection of copper resistance genes in Xanthomonas campestris pv. vesicatoria. Appl Environ Microbiol. 1991 Aug;57(8):2435–2439. doi: 10.1128/aem.57.8.2435-2439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D. C., Cody Y. S., Proebsting E. L., Radamaker G. K., Spotts R. A. Distribution, population dynamics, and characteristics of ice nucleation-active bacteria in deciduous fruit tree orchards. Appl Environ Microbiol. 1983 Dec;46(6):1370–1379. doi: 10.1128/aem.46.6.1370-1379.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S. S., Upper C. D. Dynamics, spread, and persistence of a single genotype of Pseudomonas syringae relative to those of its conspecifics on populations of snap bean leaflets. Appl Environ Microbiol. 1993 Apr;59(4):1082–1091. doi: 10.1128/aem.59.4.1082-1091.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- Kogan S. C., Doherty M., Gitschier J. An improved method for prenatal diagnosis of genetic diseases by analysis of amplified DNA sequences. Application to hemophilia A. N Engl J Med. 1987 Oct 15;317(16):985–990. doi: 10.1056/NEJM198710153171603. [DOI] [PubMed] [Google Scholar]
- Lee Y. A., Hendson M., Panopoulos N. J., Schroth M. N. Molecular cloning, chromosomal mapping, and sequence analysis of copper resistance genes from Xanthomonas campestris pv. juglandis: homology with small blue copper proteins and multicopper oxidase. J Bacteriol. 1994 Jan;176(1):173–188. doi: 10.1128/jb.176.1.173-188.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legard D. E., Aquadro C. F., Hunter J. E. DNA sequence variation and phylogenetic relationships among strains of Pseudomonas syringae pv. syringae inferred from restriction site maps and restriction fragment length polymorphism. Appl Environ Microbiol. 1993 Dec;59(12):4180–4188. doi: 10.1128/aem.59.12.4180-4188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. K., Cooksey D. A. Characterization of chromosomal homologs of the plasmid-borne copper resistance operon of Pseudomonas syringae. J Bacteriol. 1993 Jul;175(14):4492–4498. doi: 10.1128/jb.175.14.4492-4498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow S. E., Knudsen G. R., Seidler R. J., Walter M. V., Lambou V. W., Amy P. S., Schmedding D., Prince V., Hern S. Aerial Dispersal and Epiphytic Survival of Pseudomonas syringae during a Pretest for the Release of Genetically Engineered Strains into the Environment. Appl Environ Microbiol. 1988 Jun;54(6):1557–1563. doi: 10.1128/aem.54.6.1557-1563.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
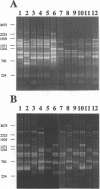
- Lupski J. R., Weinstock G. M. Short, interspersed repetitive DNA sequences in prokaryotic genomes. J Bacteriol. 1992 Jul;174(14):4525–4529. doi: 10.1128/jb.174.14.4525-4529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel M. F., Brasileiro A. C., Depierreux C., Otten L., Delmotte F., Jouanin L. Identification of different agrobacterium strains isolated from the same forest nursery. Appl Environ Microbiol. 1990 Nov;56(11):3537–3545. doi: 10.1128/aem.56.11.3537-3545.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay P., Mukhopadhyay M., Mills D. Construction of a stable shuttle vector for high-frequency transformation in Pseudomonas syringae pv. syringae. J Bacteriol. 1990 Jan;172(1):477–480. doi: 10.1128/jb.172.1.477-480.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo J., Keen N. T. Two native plasmids of Pseudomonas syringae pathovar tomato strain PT23 share a large amount of repeated DNA, including replication sequences. Mol Microbiol. 1994 Jun;12(6):941–950. doi: 10.1111/j.1365-2958.1994.tb01082.x. [DOI] [PubMed] [Google Scholar]
- Norelli J. L., Burr T. J., Lo Cicero A. M., Gilbert M. T., Katz B. H. Homologous Streptomycin Resistance Gene Present among Diverse Gram-Negative Bacteria in New York State Apple Orchards. Appl Environ Microbiol. 1991 Feb;57(2):486–491. doi: 10.1128/aem.57.2.486-491.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich J. J., Willis D. K. A single oligonucleotide can be used to rapidly isolate DNA sequences flanking a transposon Tn5 insertion by the polymerase chain reaction. Nucleic Acids Res. 1990 Nov 25;18(22):6673–6676. doi: 10.1093/nar/18.22.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield P. R., Gibson A. H., Dudman W. F., Watson J. M. Evidence for genetic exchange and recombination of Rhizobium symbiotic plasmids in a soil population. Appl Environ Microbiol. 1987 Dec;53(12):2942–2947. doi: 10.1128/aem.53.12.2942-2947.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz P., Haring V., Wittmann-Liebold B., Ashman K., Bagdasarian M., Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989 Feb 20;75(2):271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- Sundin G. W., Bender C. L. Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringae pv. syringae. Appl Environ Microbiol. 1993 Apr;59(4):1018–1024. doi: 10.1128/aem.59.4.1018-1024.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic J., Koeuth T., Lupski J. R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991 Dec 25;19(24):6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloudakis A. E., Bender C. L., Cooksey D. A. Similarity between Copper Resistance Genes from Xanthomonas campestris and Pseudomonas syringae. Appl Environ Microbiol. 1993 May;59(5):1627–1634. doi: 10.1128/aem.59.5.1627-1634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Whittam T. S., Berg C. M., Berg D. E. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993 Dec 25;21(25):5930–5933. doi: 10.1093/nar/21.25.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., Pretzman C., Postic D., Saint Girons I., Baranton G., McClelland M. Genomic fingerprinting by arbitrarily primed polymerase chain reaction resolves Borrelia burgdorferi into three distinct phyletic groups. Int J Syst Bacteriol. 1992 Jul;42(3):370–377. doi: 10.1099/00207713-42-3-370. [DOI] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F. J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992 Jul;58(7):2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]