Abstract
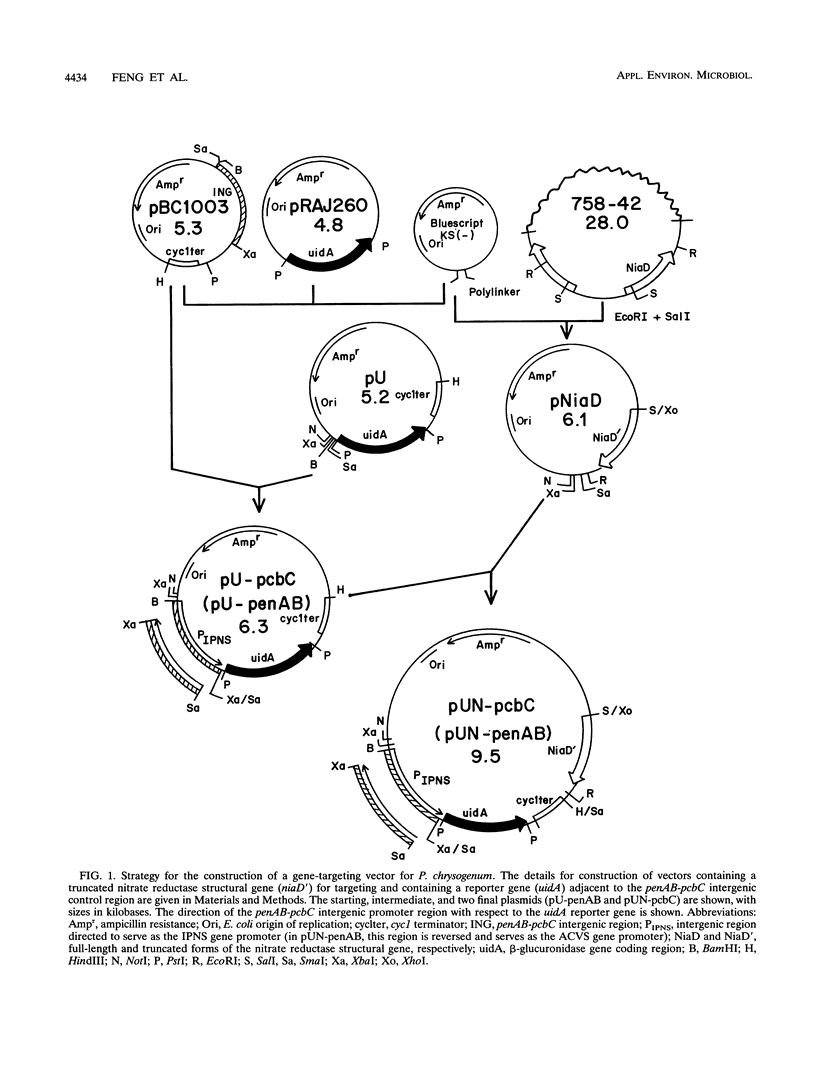
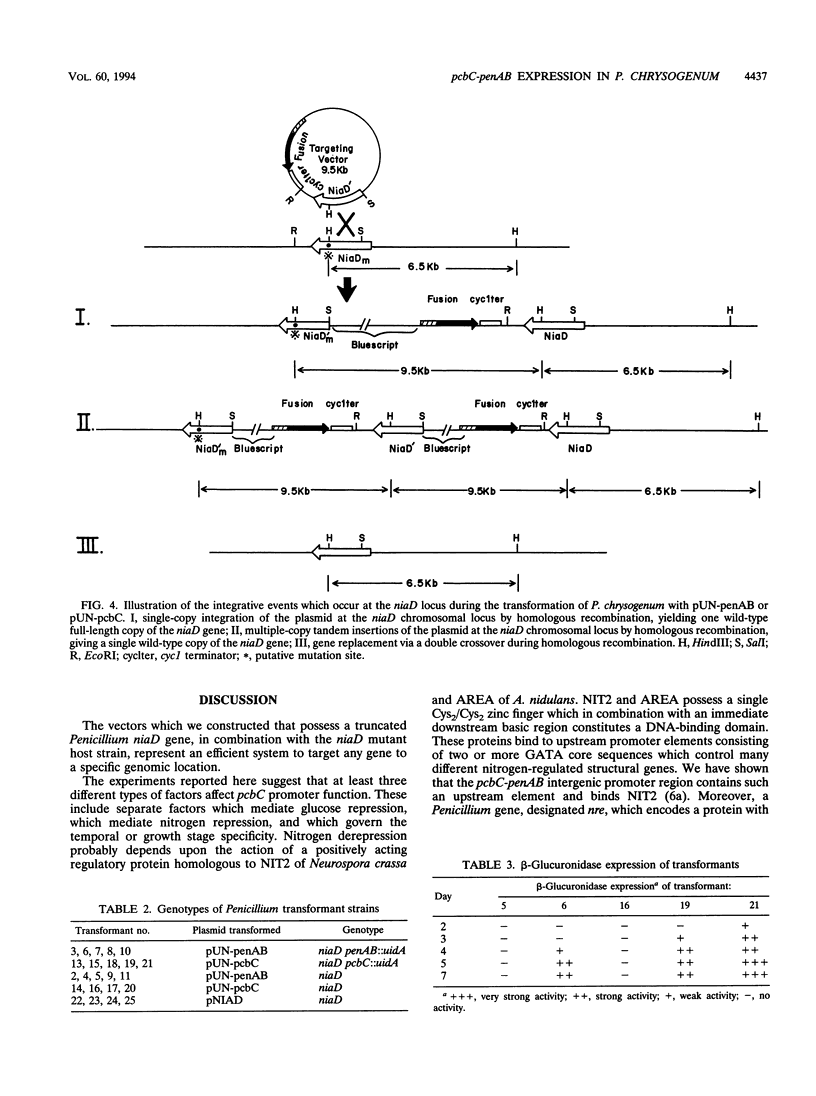
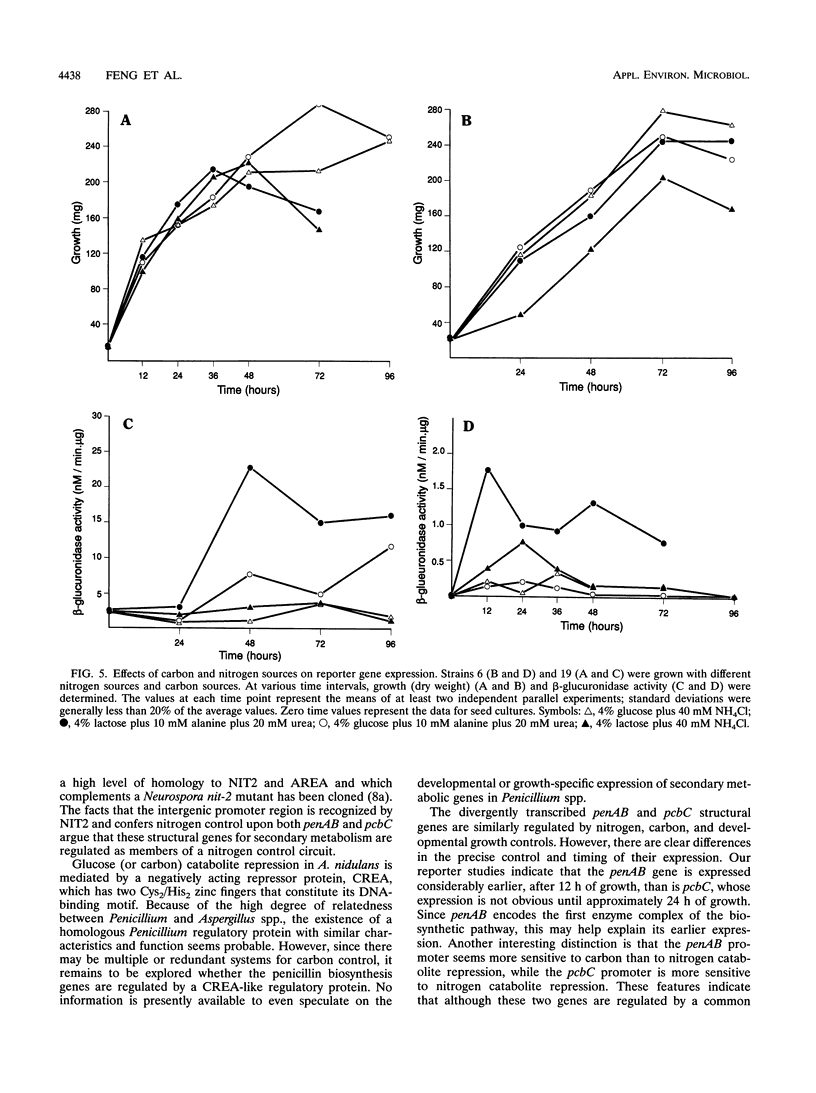
Vectors which possess a truncated niaD gene encoding nitrate reductase were developed to allow targeted gene integration during transformation of an niaD mutant Penicillium chrysogenum host. The Penicillium genes pcbC and penAB are immediately adjacent to each other and are divergently transcribed, with an intergenic control region serving as their promoters. Gene fusions were constructed with a reporter gene, uidA, which encodes beta-glucuronidase. The pcbC-penAB intergenic region was fused to the uidA gene in both orientations so that regulated expression of each structural gene could be investigated. These fusion genes were targeted to the chromosomal site of the niaD locus of P. chrysogenum, and their expression was examined under different growth conditions. The expression of each of these penicillin biosynthesis genes was found to be regulated by nitrogen repression, glucose repression, and growth stage control.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barredo J. L., Cantoral J. M., Alvarez E., Díez B., Martín J. F. Cloning, sequence analysis and transcriptional study of the isopenicillin N synthase of Penicillium chrysogenum AS-P-78. Mol Gen Genet. 1989 Mar;216(1):91–98. doi: 10.1007/BF00332235. [DOI] [PubMed] [Google Scholar]
- Brakhage A. A., Browne P., Turner G. Regulation of Aspergillus nidulans penicillin biosynthesis and penicillin biosynthesis genes acvA and ipnA by glucose. J Bacteriol. 1992 Jun;174(11):3789–3799. doi: 10.1128/jb.174.11.3789-3799.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr L. G., Skatrud P. L., Scheetz M. E., 2nd, Queener S. W., Ingolia T. D. Cloning and expression of the isopenicillin N synthetase gene from Penicillium chrysogenum. Gene. 1986;48(2-3):257–266. doi: 10.1016/0378-1119(86)90084-3. [DOI] [PubMed] [Google Scholar]
- Espeso E. A., Peñalva M. A. Carbon catabolite repression can account for the temporal pattern of expression of a penicillin biosynthetic gene in Aspergillus nidulans. Mol Microbiol. 1992 Jun;6(11):1457–1465. doi: 10.1111/j.1365-2958.1992.tb00866.x. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. Metabolic control and autogenous regulation of nit-3, the nitrate reductase structural gene of Neurospora crassa. J Bacteriol. 1988 Feb;170(2):657–661. doi: 10.1128/jb.170.2.657-661.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouka R. J., van Hartingsveldt W., Bovenberg R. A., van den Hondel C. A., van Gorcom R. F. Cloning of the nitrate-nitrite reductase gene cluster of Penicillium chrysogenum and use of the niaD gene as a homologous selection marker. J Biotechnol. 1991 Sep;20(2):189–199. doi: 10.1016/0168-1656(91)90227-m. [DOI] [PubMed] [Google Scholar]
- Jefferson R. A., Burgess S. M., Hirsh D. beta-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar M., Holzmann K., Weber G., Leitner E., Schwab H. Molecular characterization and functional analysis in Aspergillus nidulans of the 5'-region of the Penicillium chrysogenum isopenicillin N synthetase gene. J Biotechnol. 1991 Jan;17(1):67–80. doi: 10.1016/0168-1656(91)90027-s. [DOI] [PubMed] [Google Scholar]
- MacCabe A. P., Riach M. B., Unkles S. E., Kinghorn J. R. The Aspergillus nidulans npeA locus consists of three contiguous genes required for penicillin biosynthesis. EMBO J. 1990 Jan;9(1):279–287. doi: 10.1002/j.1460-2075.1990.tb08106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. R., Ingolia T. D. Cloning and characterization of beta-lactam biosynthetic genes. Mol Microbiol. 1989 May;3(5):689–695. doi: 10.1111/j.1365-2958.1989.tb00217.x. [DOI] [PubMed] [Google Scholar]
- Paietta J. V., Akins R. A., Lambowitz A. M., Marzluf G. A. Molecular cloning and characterization of the cys-3 regulatory gene of Neurospora crassa. Mol Cell Biol. 1987 Jul;7(7):2506–2511. doi: 10.1128/mcb.7.7.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Esteban B., Orejas M., Gómez-Pardo E., Peñalva M. A. Molecular characterization of a fungal secondary metabolism promoter: transcription of the Aspergillus nidulans isopenicillin N synthetase gene is modulated by upstream negative elements. Mol Microbiol. 1993 Aug;9(4):881–895. doi: 10.1111/j.1365-2958.1993.tb01746.x. [DOI] [PubMed] [Google Scholar]
- Renno D. V., Saunders G., Bull A. T., Holt G. Transcript analysis of penicillin genes from Penicillium chrysogenum. Curr Genet. 1992 Jan;21(1):49–54. doi: 10.1007/BF00318654. [DOI] [PubMed] [Google Scholar]
- Revilla G., López-Nieto M. J., Luengo J. M., Martín J. F. Carbon catabolite repression of penicillin biosynthesis by Penicillium chrysogenum. J Antibiot (Tokyo) 1984 Jul;37(7):781–789. doi: 10.7164/antibiotics.37.781. [DOI] [PubMed] [Google Scholar]
- Smith D. J., Burnham M. K., Bull J. H., Hodgson J. E., Ward J. M., Browne P., Brown J., Barton B., Earl A. J., Turner G. Beta-lactam antibiotic biosynthetic genes have been conserved in clusters in prokaryotes and eukaryotes. EMBO J. 1990 Mar;9(3):741–747. doi: 10.1002/j.1460-2075.1990.tb08168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff H. B. Natural products from microorganisms. Science. 1980 Jun 13;208(4449):1225–1229. doi: 10.1126/science.7375932. [DOI] [PubMed] [Google Scholar]




