Abstract
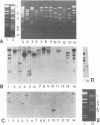
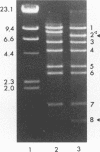
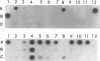
In the last 30 years, 81 Streptococcus thermophilus bacteriophage isolates were collected from industrial yogurt (n = 40) and cheese (n = 41) fermentation. Forty-six distinct restriction patterns of phage DNA (11 in yogurt and 35 in cheese) were observed. The phages were investigated for host range, serological properties, and DNA homology to study whether these three independent techniques can be used to classify the phages into taxonomic groups. Yogurt factory-derived phages were classified into the same two subgroups by serology, host range analysis, and hybridization with subgroup-specific DNA sequences. Cheese factory-derived phages, however, could not be classified: the 35 cheese phage isolates with distinct restriction patterns showed 34 different host ranges. All but one cheese phage isolate showed serological cross-reactivity with yogurt phages. A phage DNA fragment that hybridized with all phage DNA samples was cloned, establishing the genetic relatedness of all S. thermophilus phages from our collection. With the sequence information from an unusually conserved S. thermophilus phage DNA element (H. Brüssow, A. Probst, M. Frémont, and J. Sidoti, Virology 200:854-857, 1994), a PCR-based phage detection method was developed for cheese whey from a factory that produced mozzarella cheese with complex undefined starter mixes. PCR allowed the detection of phages in cheese whey (detection limit, 10(3) PFU/ml) which could not be detected by dot blot hybridization techniques (detection limit, 10(7) PFU/ml).
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benbadis L., Faelen M., Slos P., Fazel A., Mercenier A. Characterization and comparison of virulent bacteriophages of Streptococcus thermophilus isolated from yogurt. Biochimie. 1990 Dec;72(12):855–862. doi: 10.1016/0300-9084(90)90002-x. [DOI] [PubMed] [Google Scholar]
- Bradley D. E. Ultrastructure of bacteriophage and bacteriocins. Bacteriol Rev. 1967 Dec;31(4):230–314. doi: 10.1128/br.31.4.230-314.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüssow H., Probst A., Frémont M., Sidoti J. Distinct Streptococcus thermophilus bacteriophages share an extremely conserved DNA fragment. Virology. 1994 May 1;200(2):854–857. doi: 10.1006/viro.1994.1256. [DOI] [PubMed] [Google Scholar]
- Haggård-Ljungquist E., Halling C., Calendar R. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J Bacteriol. 1992 Mar;174(5):1462–1477. doi: 10.1128/jb.174.5.1462-1477.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highton P. J., Chang Y., Myers R. J. Evidence for the exchange of segments between genomes during the evolution of lambdoid bacteriophages. Mol Microbiol. 1990 Aug;4(8):1329–1340. doi: 10.1111/j.1365-2958.1990.tb00712.x. [DOI] [PubMed] [Google Scholar]
- Kim S. G., Batt C. A. Identification of a nucleotide sequence conserved in Lactococcus lactis bacteriophages. Gene. 1991 Feb 1;98(1):95–100. doi: 10.1016/0378-1119(91)90109-o. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larbi D., Decaris B., Simonet J. M. Different bacteriophage resistance mechanisms in Streptococcus salivarius subsp. thermophilus. J Dairy Res. 1992 Aug;59(3):349–357. doi: 10.1017/s0022029900030624. [DOI] [PubMed] [Google Scholar]
- Moineau S., Bernier D., Jobin M., Hébert J., Klaenhammer T. R., Pandian S. Production of Monoclonal Antibodies against the Major Capsid Protein of the Lactococcus Bacteriophage ul36 and Development of an Enzyme-Linked Immunosorbent Assay for Direct Phage Detection in Whey and Milk. Appl Environ Microbiol. 1993 Jul;59(7):2034–2040. doi: 10.1128/aem.59.7.2034-2040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. E., Summers M. D. The bidirectional transfer of DNA and RNA to nitrocellulose or diazobenzyloxymethyl-paper. Anal Biochem. 1980 Nov 15;109(1):123–129. doi: 10.1016/0003-2697(80)90019-6. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasala A., Dupont L., Baumann M., Ritzenthaler P., Alatossava T. Molecular comparison of the structural proteins encoding gene clusters of two related Lactobacillus delbrueckii bacteriophages. J Virol. 1993 Jun;67(6):3061–3068. doi: 10.1128/jvi.67.6.3061-3068.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhue W. M. Interaction of bacteriophage infection and low penicillin concentrations on the performance of yogurt cultures. Appl Environ Microbiol. 1978 Jun;35(6):1145–1149. doi: 10.1128/aem.35.6.1145-1149.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernars K., Heuvelman C. J., Chakraborty T., Notermans S. H. Use of the polymerase chain reaction for direct detection of Listeria monocytogenes in soft cheese. J Appl Bacteriol. 1991 Feb;70(2):121–126. doi: 10.1111/j.1365-2672.1991.tb04437.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]