Abstract
Enzymes with high specific activities at low temperatures have potential uses for chemical conversions when low temperatures are required, as in the food industry. Psychrotrophic microorganisms which grow at low temperatures may be a valuable source of cold-active enzymes that have higher activities at low temperatures than enzymes found for mesophilic microorganisms. To find cold-active beta-galactosidases, we isolated and characterized several psychrotrophic microorganisms. One isolate, B7, is an Arthrobacter strain which produces beta-galactosidase when grown in lactose minimal media. Extracts have a specific activity at 30 degrees C of 2 U/mg with o-nitrophenyl-beta-D-galactopyranoside as a substrate. Two isozymes were detected when extracts were subjected to electrophoresis in a nondenaturing polyacrylamide gel and stained for activity with 5-bromo-4-chloro-indolyl-beta-D-galactopyranoside (X-Gal). When chromosomal DNA was prepared and transformed into Escherichia coli, three different genes encoding beta-galactosidase activity were obtained. We have subcloned and sequenced one of these beta-galactosidase genes from the Arthrobacter isolate B7. On the basis of amino acid sequence alignment, the gene was found to have probable catalytic sites homologous to those from the E. coli lacZ gene. The gene encoded a protein of 1,016 amino acids with a predicted molecular mass of 111 kDa. The enzyme was purified and characterized. The beta-galactosidase from isolate B7 has kinetic properties similar to those of the E. coli lacZ beta-galactosidase but has a temperature optimum 20 degrees C lower than that of the E. coli enzyme.
Full text
PDF
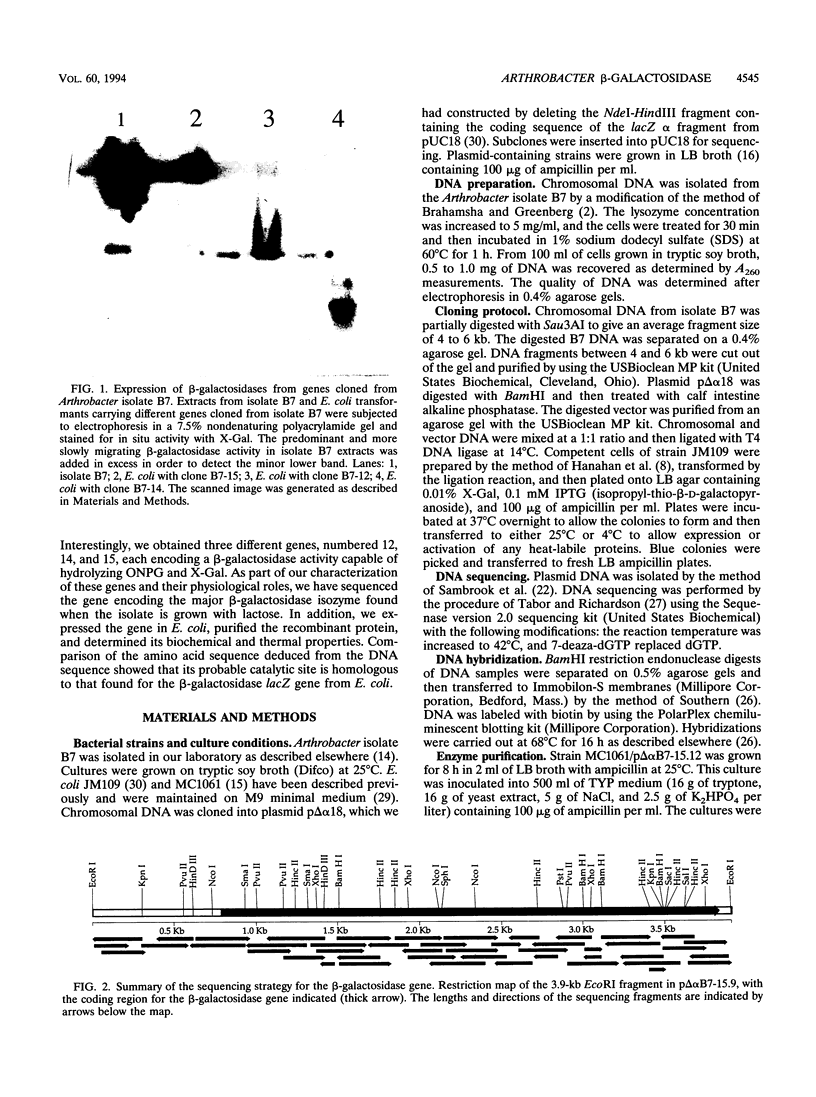





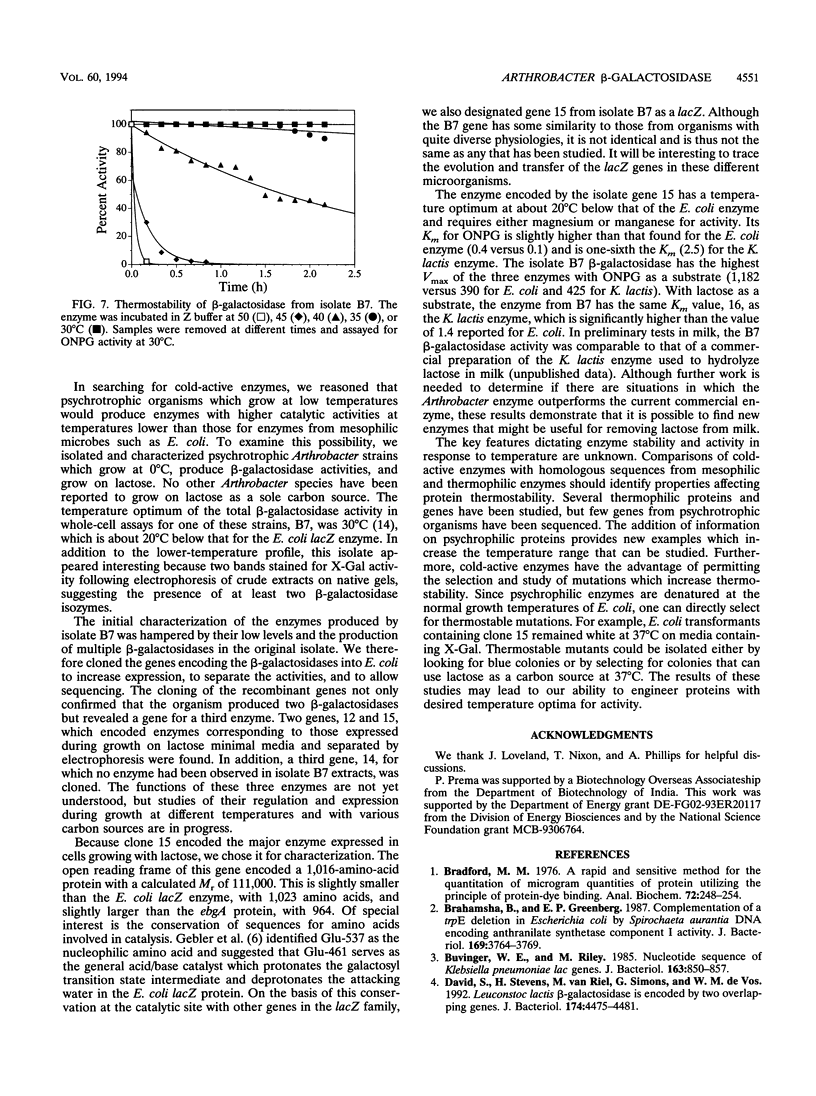

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brahamsha B., Greenberg E. P. Complementation of a trpE deletion in Escherichia coli by Spirochaeta aurantia DNA encoding anthranilate synthetase component I activity. J Bacteriol. 1987 Aug;169(8):3764–3769. doi: 10.1128/jb.169.8.3764-3769.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvinger W. E., Riley M. Nucleotide sequence of Klebsiella pneumoniae lac genes. J Bacteriol. 1985 Sep;163(3):850–857. doi: 10.1128/jb.163.3.850-857.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David S., Stevens H., van Riel M., Simons G., de Vos W. M. Leuconostoc lactis beta-galactosidase is encoded by two overlapping genes. J Bacteriol. 1992 Jul;174(13):4475–4481. doi: 10.1128/jb.174.13.4475-4481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Dickson L. R., Markin J. S. Purification and properties of an inducible beta-galactosidase isolated from the yeast Kluyveromyces lactis. J Bacteriol. 1979 Jan;137(1):51–61. doi: 10.1128/jb.137.1.51-61.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L. In vivo mutagenesis. Methods Enzymol. 1991;204:114–125. doi: 10.1016/0076-6879(91)04007-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebler J. C., Aebersold R., Withers S. G. Glu-537, not Glu-461, is the nucleophile in the active site of (lac Z) beta-galactosidase from Escherichia coli. J Biol Chem. 1992 Jun 5;267(16):11126–11130. [PubMed] [Google Scholar]
- Hall B. G., Betts P. W., Wootton J. C. DNA sequence analysis of artificially evolved ebg enzyme and ebg repressor genes. Genetics. 1989 Dec;123(4):635–648. doi: 10.1093/genetics/123.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock K. R., Rockman E., Young C. A., Pearce L., Maddox I. S., Scott D. B. Expression and nucleotide sequence of the Clostridium acetobutylicum beta-galactosidase gene cloned in Escherichia coli. J Bacteriol. 1991 May;173(10):3084–3095. doi: 10.1128/jb.173.10.3084-3095.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrchen M., Legler G. Identification of an essential carboxylate group at the active site of lacZ beta-galactosidase from Escherichia coli. Eur J Biochem. 1984 Feb 1;138(3):527–531. doi: 10.1111/j.1432-1033.1984.tb07947.x. [DOI] [PubMed] [Google Scholar]
- Kalnins A., Otto K., Rüther U., Müller-Hill B. Sequence of the lacZ gene of Escherichia coli. EMBO J. 1983;2(4):593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leary J. J., Brigati D. J., Ward D. C. Rapid and sensitive colorimetric method for visualizing biotin-labeled DNA probes hybridized to DNA or RNA immobilized on nitrocellulose: Bio-blots. Proc Natl Acad Sci U S A. 1983 Jul;80(13):4045–4049. doi: 10.1073/pnas.80.13.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland J., Gutshall K., Kasmir J., Prema P., Brenchley J. E. Characterization of psychrotrophic microorganisms producing beta-galactosidase activities. Appl Environ Microbiol. 1994 Jan;60(1):12–18. doi: 10.1128/aem.60.1.12-18.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner P. S., Sisk W. P., Berman M. L. Bacteriophage lambda cloning system for the construction of directional cDNA libraries. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4171–4175. doi: 10.1073/pnas.84.12.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naider F., Bohak Z., Yariv J. Reversible alkylation of a methionyl residue near the active site of -galactosidase. Biochemistry. 1972 Aug 15;11(17):3202–3208. doi: 10.1021/bi00767a010. [DOI] [PubMed] [Google Scholar]
- Poch O., L'Hôte H., Dallery V., Debeaux F., Fleer R., Sodoyer R. Sequence of the Kluyveromyces lactis beta-galactosidase: comparison with prokaryotic enzymes and secondary structure analysis. Gene. 1992 Sep 1;118(1):55–63. doi: 10.1016/0378-1119(92)90248-n. [DOI] [PubMed] [Google Scholar]
- Saavedra J. M., Perman J. A. Current concepts in lactose malabsorption and intolerance. Annu Rev Nutr. 1989;9:475–502. doi: 10.1146/annurev.nu.09.070189.002355. [DOI] [PubMed] [Google Scholar]
- Schachter H. Enzymic microassays for D-mannose, D-glucose, D-galactose, L-fucose, and D-glucosamine. Methods Enzymol. 1975;41:3–10. doi: 10.1016/s0076-6879(75)41003-5. [DOI] [PubMed] [Google Scholar]
- Schmidt B. F., Adams R. M., Requadt C., Power S., Mainzer S. E. Expression and nucleotide sequence of the Lactobacillus bulgaricus beta-galactosidase gene cloned in Escherichia coli. J Bacteriol. 1989 Feb;171(2):625–635. doi: 10.1128/jb.171.2.625-635.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder C. J., Robert C., Lenzen G., McKay L. L., Mercenier A. Analysis of the lacZ sequences from two Streptococcus thermophilus strains: comparison with the Escherichia coli and Lactobacillus bulgaricus beta-galactosidase sequences. J Gen Microbiol. 1991 Feb;137(2):369–380. doi: 10.1099/00221287-137-2-369. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]