Abstract
Heme proteins play pivotal roles in a wealth of biological processes. Despite this, the molecular mechanisms by which heme traverses bilayer membranes for use in biosynthetic reactions are unknown. The biosynthesis of c-type cytochromes requires that heme is transported to the bacterial periplasm or mitochondrial intermembrane space where it is covalently ligated to two reduced cysteinyl residues of the apocytochrome. Results herein suggest that a family of integral membrane proteins in prokaryotes, protozoans, and plants act as transmembrane heme delivery systems for the biogenesis of c-type cytochromes. The complete topology of a representative from each of the three subfamilies was experimentally determined. Key histidinyl residues and a conserved tryptophan-rich region (designated the WWD domain) are positioned at the site of cytochrome c assembly for all three subfamilies. These histidinyl residues were shown to be essential for function in one of the subfamilies, an ABC transporter encoded by helABCD. We believe that a directed heme delivery pathway is vital for the synthesis of cytochromes c, whereby heme iron is protected from oxidation via ligation to histidinyl residues within the delivery proteins.
The structures and functions of heme proteins, and in some cases their assembly pathways, are well characterized (e.g., refs. 1–3). These proteins are fundamental in processes ranging from energy conversion to detoxification to oxygen transport. Nevertheless, the mechanisms by which heme is delivered intracellularly to different sites for the assembly of heme proteins are poorly understood. Specific sites of synthesis in eukaryotes include the cytoplasm (e.g., hemoglobin), mitochondrial matrix (e.g., cytochrome b), endoplasmic reticulum (e.g., cytochrome P450), and the mitochondrial intermembrane space (e.g., cytochrome c). In prokaryotic cells, heme proteins are synthesized in the cytoplasm or inner (cytoplasmic) membrane (e.g., cytochrome oxidase) or outside the inner membrane (e.g., cytochrome c). Although it has been commonly assumed that free heme diffuses throughout the cell, this assumption may not be true for sites where heme could catalyze the formation of damaging reactive oxygen species (e.g., membranes).
Another example where oxidized heme is detrimental concerns the biogenesis of c-type cytochromes. Transport of heme to the bacterial periplasm (e.g., refs. 4 and 5) or mitochondrial intermembrane space (e.g., refs. 6 and 7) is required for the biosynthesis of c-type cytochromes. The iron of heme must also be reduced (8, 9) for the covalent attachment of heme vinyl groups to two reduced cysteinyl residues of the apocytochrome c (10–13). Such considerations raise the possibility that transmembrane heme delivery systems may be required by organisms for specific biosynthetic processes. We provide herein evidence that a family of proteins in a wide variety of prokaryotes and eukaryotes may function as heme delivery systems for the biogenesis of c-type cytochromes.
From their amino acid sequences, we have described (4) a family of proteins distinguished by a conserved tryptophan-rich region (designated here the WWD domain). By analyses of the genomic databases, the following three subfamilies were identified: (i) the HelC protein, a predicted membrane subunit of an ATP-binding cassette (ABC) transporter encoded by the helABCD genes of Gram-negative bacteria; (ii) a 653-residue protein called Ccl1, also in Gram-negative bacteria such as Rhodobacter capsulatus and Escherichia coli; and (iii) the 327-residue CcsA protein of chloroplasts. Genetic analysis in bacteria have established that the helABCD and the ccl1 genes are necessary in some Gram-negative bacteria for cytochrome c biogenesis (4, 5, 11–16). The HelA protein is the ATP-binding subunit of the HelABCD transporter, and we have demonstrated that HelA requires the two membrane components, HelB and HelC, for formation of a four-subunit macromolecular export complex (5). Recent genomic analyses indicate that genes encoding the HelABC and Ccl1 proteins are also present in plant and protozoal mitochondria (17–20). The CcsA protein is encoded in the genomes of chloroplasts and genetic analysis in Chlamydomonas indicates that the ccsA gene is necessary for cytochrome c biogenesis in the chloroplast (21). The ccsA gene is also in Gram-positive bacteria such as Mycobacterium leprae (GenBank accession no. U00018) and Bacillus subtilis (22). In spite of the widespread occurrence of this family of proteins, very little is known about their exact functions or structures. The present study was undertaken to establish the transmembrane topology of all three members of the family. Additionally, the functional significance of key residues in the HelABCD exporter was determined. These residues are analogous to important residues for heme binding in other proteins, including hemoglobin (23) and hemopexin (24).
MATERIALS AND METHODS
Strains and Plasmids.
Plasmids containing HelB, HelC, and Ccl1 alkaline phosphatase (phoA) translational fusions were constructed as described (4). All oligonucleotides used for generating fusions in this study and the color figures can be found on the internet (biosgi.wustl.edu/faculty/kranz.html). The hel or ccl1 genes were amplified by the PCR using the M13 reverse primer and a synthetic oligonucleotide primer designed to engineer a SalI site at the desired fusion junction (see Fig. 1 for locations of the junctions). The Klentaq LA (long and accurate) polymerase mixture (CLONTECH) was used for all PCRs due to its extremely high fidelity. Twenty-five cycles were used to generate all PCR products. Given an error frequency of <10−6 (25), there is a <5% chance of an introduced DNA error for the ccl1 genes and a <2.5% chance of an introduced DNA error for the ccsA, helB, and helC genes. These are the error maxima, so that the likelihood of an actual error is probably considerably less (25). The plasmids pRGK201 (5) and pRGK203 (16) were used as template for hel and ccl1 PCRs, respectively. The amplified hel products were cloned into pRGK255 containing phoA (minus its signal sequence) as SalI–KpnI fragments. The amplified ccl1 product with the C3 domain as its C terminus was cloned into pUC118 with KpnI and SalI. phoA from pRGK200 was then cloned into this plasmid (pRGK298) at SalI–HindIII restriction sites. Other amplified ccl1 products were cloned into pRGK298 at the KpnI–SalI restriction sites. The lacZ fusion vectors pLKC480 and pLKC482 were provided by John Smith (Seattle Biomedical Research Institute) (26). The C6 (C terminus) Ccl1-LacZ translational fusion was created by using the 6.3-kb SalI–SmaI fragment containing the lacZ gene from pLKC480, whereas the 3Pa, 3Pb, and C3 Ccl1-LacZ fusions contain the SalI–SmaI fragment from pLKC482. These fusion constructs were made in two steps. First, each pUC118 construct containing the amplified DNA insert was digested with SphI, the ends were filled by using the Klenow fragment of DNA polymerase I, and subsequently digested with SalI. The SalI–SmaI fragment containing the lacZ gene (and kanR gene) was ligated into these sites. The ccl-lacZ constructs were digested with KpnI and cloned into the broad host vector pUCA10. The C termini of the HelB, HelC, and HelD proteins have been shown to be located in the cytoplasm (5). E. coli CC118 (Lac− Pho−) was transformed with all phoA fusions plasmids and used for alkaline phosphatase analysis. Bacterial colonies were also screened for alkaline phosphatase by using 5-bromo-4-chloro-3-indolyl phosphate (Sigma), as described by Manoil and Beckwith (27). These plate assays were consistent with activity measurements (Table 1). For ccsA-phoA gene fusions, a BamHI–XhoI fragment containing the phoA gene from plasmid pRGK200 was cloned into pET21b at the BamHI–XhoI site to generate plasmid pETphoA. The ccsA gene from M. leprae was amplified from genomic DNA (provided by J. Clark-Curtiss, Washington University, St. Louis, MO) by the PCR using the oligonucleotides 5′-GAACGTCATATGAACACTCTGCAGGTCAACATC-3′ (upstream) and 5′-GGTCTGCATTCGTATGCGGAAGTGGAAGATCTCGAGGTT-3′ (downstream). These oligonucleotides generated an NdeI site upstream of the gene and BglII and XhoI sites downstream. pETCcsA was made by cloning the NdeI–XhoI fragment of the amplified ccsA gene into the plasmid pET21b at the NdeI–XhoI sites. pETCcsA was used as the template for the phoA fusions. The ccsA PCR fragments for each fusion were digested with NdeI–BglII and cloned into the pETPhoA vector at the NdeI–BamHI restriction sites. These phoA fusion plasmids were transformed into the E. coli BL21/DE3 for alkaline phosphatase analysis. The ccsA, helB, and helC junctions were sequenced and each fusion junction was in-frame and no changes observed spanning at least 100 nucleotides of the junctions. The presence of CcsA-PhoA fusion proteins were confirmed by Western blot analysis using antibodies to alkaline phosphatase (5 Prime–3 Prime, Inc.).
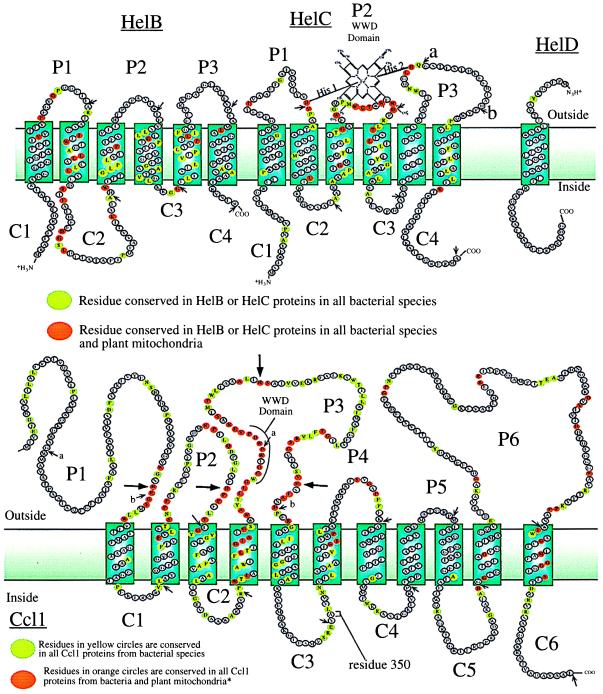
Figure 1.
Topology of proteins involved in heme delivery for cytochrome c biogenesis in Gram-negative bacteria and plant mitochondria. The indicated sequences are from R. capsulatus proteins. For the HelC and Ccl1 proteins, the conserved tryptophan-rich region (called the WWD domain) is periplasmic and is flanked by histidinyl residues (large arrows). (Upper) The topology of the integral membrane proteins of the HelABCD transporter were determined by the analysis of PhoA fusion proteins (see Table 1) that were created at each hydrophilic domain (shown by small arrows). Conserved residues within the bacterial proteins are from E. coli, Haemophilus influenzae, Paracoccus denitrificans, Pseudomonas fluorescens, and Bradyrhizobium japonicum. For conserved residues between the HelC proteins of bacteria and plant mitochondria, the above organisms and Oenothera, Dauca, and Marchantia were used. For conserved residues between the HelB proteins of bacteria and plant mitochondria compared the above organisms and Marchantia were used. The heme iron is coordinated to the His-1 and His-2 residues of the HelC protein, as discussed in text. The 52-residue HelD polypeptide was shown to have its C terminus in the cytoplasm. (Lower) The topology of the integral membrane heme protein Ccl1 was determined by analysis of PhoA fusion proteins (Table 1) that were created at each hydrophilic domain (shown by small arrows). Conserved residues of Ccl1 between bacteria compared the proteins from R. capsulatus, E. coli, H. influenzae, P. denitrificans, Rhizobium meliloti, and B. japonicum. Residues from 1 to 350 of the bacterial Ccl1 protein that are also conserved with the plant mitochondria ORF577 of Oenothera, Dauca, and Marchantia. Residues from 351 to 651 are from ORF454 of Oenothera, Brassica, and Arabadopsis.
Table 1.
Alkaline phosphatase activities of PhoA fusions to members of the three subfamilies of transmembrane heme delivery proteins
| Hel fusion | Specific activity, units | Ccl1 fusion | Specific activity, units | CcsA fusion | Specific activity, units |
|---|---|---|---|---|---|
| HelB P1 | 340 | P1a | 220 | C1a | 24 |
| HelB C2 | 8 | P1b (His-1) | 350 | C1b | 17 |
| HelB P2 | 130 | C1 | 14 | P2a | 680 |
| HelB C3 | <1 | P2 (His-2) | 320 | P2b (His-1) | ND* |
| HelB P3 | 100 | C2 | 5 | C2 | 26 |
| HelB C4 C terminus | 6 | P3a (WWD, His-3) | 360 | P3 (WWD) | 170 |
| P3b (His-4) | 880 | C3 | 24 | ||
| HelC P1 | 130 | C3 | 30 | P4 C terminus (His-2) | 260 |
| HelC C2 | 16 | P4 | 260 | pET-phoA vector in BL21 | 8 |
| HelC P2 (His-1) | 180 | C4 | <1 | ||
| HelC C3 | 6 | P5 | 490 | BL21 | 3 |
| HelC P3a (His-2) | 40 | C5 | <1 | ||
| HelC P3b | 22 | P6 | 390 | ||
| HelC4 C terminus | <1 | C6 C terminus | 15 | ||
| CC118 | <1 | CC118 | <1 |
Fusions to periplasmic domains are called P and those to the cytoplasmic domains are called C as depicted in Figs. 1 and 2. Alkaline phosphatase activities were obtained in E. coli; results are consistent with alkaline phosphatase assays performed with several of the above fusions in R. capsulatus.
The specific activity of this strain could not be determined (ND) due to the toxic effect of the fusion protein on the cells upon induction by isopropyl β-d-thiogalactoside. However, like other PhoA fusion proteins that have periplasmic orientations, the fusion protein at P2b was not degraded, as determined by Western blot analysis. In contrast, the cytoplasmic fusion proteins were unstable and degraded (data not shown).
Enzyme Assays.
For alkaline phosphatase and β-galactosidase assays in E. coli, cells were grown aerobically at 37°C in LB broth supplemented with ampicillin (200 μg/ml) for plasmid selection until early exponential phase. The cultures were then induced with 0.3 mM isopropyl β-d-thiogalactopyranoside for 2 h. Whole cells were used in the alkaline phosphatase assays and sonicated cells were used for β-galactosidase assays. Results are averages from at least three experiments each done in triplicate with each assay less than 2-fold different from the average.
Site-Directed Mutagenesis.
The helC and ccl1 genes were mutagenized on pUC118 plasmids containing the helBCDX and ccl12 genes, respectively, by the method of Kunkel (28) using the Muta-Gene kit (Bio-Rad). After the base alterations were confirmed by sequence analysis, the pUC118 plasmids containing the mutated and wild-type helC and ccl1 genes were cloned into the pUCA10 plasmid at the HindIII site. The resulting ampicillin-resistant tetracycline-resistant plasmids were conjugated into the R. capsulatus ΔhelC (5) or Δccl1 (16) strains.
RESULTS
Topology of the ABC Transporter HelABCD.
To determine the complete topology of each member of the WWD domain family of proteins, phoA fusions were engineered in-frame to all of the predicted soluble domains in the membrane proteins HelB, HelC, Ccl1, and CcsA (Figs. 1 and 2A). In bacteria, alkaline phosphatase, encoded by the phoA gene, is often used as a topological reporter for membrane proteins. Because alkaline phosphatase only folds properly outside the membrane, high alkaline phosphatase activity indicates an external location and low activity indicates an internal location (27). The helB, helC, and ccl1 genes are from the Gram-negative photosynthetic bacterium R. capsulatus and the ccsA gene is from M. leprae. Determination of the alkaline phosphatase activities (Table 1) of cells containing each fusion protein established the topological location of each domain. For example, the highly conserved WWD domain in the HelC protein is present in the periplasm (P2) because a PhoA fusion to it generates a protein with high alkaline phosphatase activity (180 units). In contrast, the C2 domain of the HelB protein is on the cytoplasmic side because this fusion exhibited low activity (8 units). For the HelB protein, all domains designated as periplasmic generated fusions with activities at least 21-fold higher than the average activity of the cytoplasmic fusions. All periplasmic fusions in the HelC protein, except HelC P3, exhibit at least 17-fold higher activities than the average of activities for all the cytoplasmic fusions. For the HelC P3 domain, both fusions were significantly higher (40 and 22 units) than the adjacent cytoplasmic domains (6 and <1 units) with P3a having 5-fold higher activity than the average cytoplasmic fusion activity. Additionally, all colonies with plasmids containing periplasmic fusions, including P3a, were dark blue on 5-bromo-4-chloro-3-indolyl phosphate indicator plates, whereas colonies containing cytoplasmic fusions were white or light blue. Within the HelC protein, two periplasmic domains (P1 and P3) contain conserved histidinyl residues that are proximal and distal to the WWD domain (H58 and H183, referred to as His1 and His2). Results on the HelB and HelC fusion proteins (Table 1) indicate that six transmembrane domains are present in each (see Fig. 1), as is the case with other bacterial membrane transporters (for review, see ref. 30).
Figure 2.
(A) Topology of the CcsA protein of M. leprae. Residues of the CcsA protein from M. leprae, Mycobacterium tuberculosis, and B. subtilis are compared. Topology was determined by the analysis of PhoA fusion proteins (Table 1) that were created at each of the hydrophilic domains (shown by small arrows). The conserved WWD domain is external and is flanked by histidinyl residues (large arrows). For conserved residues between the CcsA protein of bacteria and chloroplasts, the above organisms and Chlamydomonas, Porphyra, and Marchantia were used. (B) Model for cytochrome c biogenesis pathway in Gram-negative bacteria, plant and protozoal mitochondria, and possibly archae, as described in the text. The apocytochrome is exported to the periplasm via the Sec-dependent pathway. In the periplasm, the signal sequence is cleaved and an intramolecular disulfide bound is catalyzed by the DsbA protein. Before ligation, the cysteinyl groups of the apocytochrome are reduced by the Ccl2 protein, which is, in turn, rereduced by the HelX protein (see ref. 12 and references therein). The CycH protein may act as a chaperone of the apocytochrome during the ligation process (see refs. 29 and 40). Heme is exported from the cytoplasm through the HelABCD transporter and, presumably, brought to the Ccl1 protein for eventual ligation to the apocytochrome.
Topology of the Ccl1 Protein.
The results of alkaline phosphatase activities of Ccl1 fusion proteins are shown in Table 1. As a test for certain domains and the PhoA reporter in general, β-Galactosidase (LacZ) fusions were also generated to the Ccl1 protein at the P3a, P3b, C3, and C6 domains. High levels of β-galactosidase activity are indicative of an internal location, whereas low activities are indicative of an external location (31). β-Galactosidase activities at domains P3a (170 units) and P3b (200 units) were low and at C3 (1,100 units) and C6 (2,500 units) were high, consistent with the PhoA analysis of these domains. Alkaline phosphatase activities of strains containing periplasmic fusions were at least 20-fold higher than the average activity of the cytoplasmic fusions. On the basis of our phoA and lacZ fusion results, the Ccl1 protein contains 11 transmembrane domains (Fig. 1). This protein is split into two ORFs in plant mitochondria, with the N-terminal half (ORF577) containing the most highly conserved residues, including the WWD domain (32). Interestingly, all residues that are conserved between the prokaryotic Ccl1 protein and eukaryotic ORF577 proteins are located in the large periplasmic domains P1, P2, and P3 (Fig. 1). These include four conserved histidinyl residues (His-92, His-172, His-260, and His-302) flanking the WWD domain. The implications for these results are discussed below.
Topology of the CcsA Protein.
To establish the topology of the third member of the family, the M. leprae ccsA gene was isolated by using PCR and expressed in E. coli as the designated fusion proteins (Fig. 2A and Table 1). A new inducible phoA reporter plasmid, called pETphoA, was constructed for this analysis. Each of the designated CcsA periplasmic fusion was at least 7-fold higher than the average cytoplasmic fusion. Although the CcsA protein clearly defines a separate member of the family, the topological results demonstrate that two histidinyl residues (His-176 and His-321) also flank the WWD domain at the periplasmic surface. The CcsA protein has six transmembrane regions with many conserved residues in both cytoplasmic and periplasmic domains. Previously, it has been reported that the chloroplast CcsA protein is soluble, as determined by membrane fractionation and detection with a peptide antibody to CcsA (21). Nevertheless, the order of predicted transmembrane domains in the chloroplast CcsA protein is similar to that in the M. leprae CcsA protein (data not shown). We confirmed that each of the M. leprae CcsA Pho fusions was membrane-bound by analyzing activities in crude and membrane fractions. These results and Western blots with alkaline phosphatase antibodies clearly demonstrated that each fusion is membrane-bound (data not shown).
Conserved Periplasmic Histidinyl Residues Flanking the WWD Domain Are Required.
From the topological profiles determined herein, we hypothesize that proteins of this family use two histidinyl residues that are proximal and distal to the WWD domain to ligand heme at the outer surface of the inner membrane. In this scenario, heme is delivered to the site of cytochrome c assembly, the bacterial periplasm, where it is coordinated by these histidinyl residues (see Fig. 2B and Discussion).
To test the requirements for residues proposed to play key roles in heme delivery and presentation, specific amino acids in the HelC protein were altered. A deficiency in cytochrome c biogenesis in R. capsulatus results in an inability to grow under anaerobic photosynthetic conditions (14). This phenotype was used to test the functions of mutated helC genes at the two histidines or within the WWD domain. The His-58 residue of the HelC protein was changed to glycine, methionine, valine, threonine, or serine. The His-183 residue was changed to methionine, cysteine, or glycine. All changes resulted in a defective transporter (see Fig. 3, for examples). To determine whether other residues at His-58 might be functional (or whether other changes could correct the HelC H58M defect) we selected revertants of the H58M mutants under photosynthetic conditions. Although rare (<10−9), three revertants were isolated. Each of the three reversions were associated with the helC gene on the plasmid. Sequence analysis of these three genes indicated that each had regained histidine at residue 58.
Figure 3.
Functional analysis of key residues in the HelC and Ccl1 proteins. (Upper) Growth of strains containing HelC WWD, histidine mutations, or Ccl1 WWD mutations under aerobic and anaerobic conditions. Under anaerobic (photosynthetic) conditions, c-type cytochromes, and thus the biogenesis proteins, are required for growth. Plasmids containing either the helC or the ccl1 genes were conjugated into a strain containing a nonpolar deletion of the helC or ccl1 gene, respectively. All strains grow aerobically due to the existence in R. capsulatus of a cytochrome c-independent electron transport pathway (14). (Lower) The WWD domain and flanking histidinyl residues are shown for all three members of the family. +, Residue(s) can be changed and the protein is still functional. Altering His-1 and His-2 of HelC to glycine generates a nonfunctional protein.
To determine whether the His-58 or His-183 mutations affected the stability of the HelABC transporter complex, the mutants containing the most radical alterations (H58G and H183G) were investigated further. We have previously shown that in strains defective for either of the integral membrane proteins (HelB or HelC), the HelA subunit is degraded and cannot be detected (5). Fig. 4 demonstrates that the HelA protein is absent in an ΔhelC mutant but is easily detected in both helC histidine mutants. Thus, both of the HelC histidinyl residues (H58G, H183G) are essential for transporter function but not for assembly of the HelABC(D) complex.
Figure 4.
Stability of the HelABC complex in HelC mutant strains. The stability of the HelABC complex was determined by analyzing the presence of the HelA protein by immunoblot of R. capsulatus ΔhelC strains containing plasmids carrying either the wild type or mutated helC gene. Lanes: 1, 2, and 4–8, equal amounts of membrane fractions (∼40 μg of protein per lane); 3, ∼20 μg of protein per lane of membrane fraction. The fractions were separated by PAGE on a 15% gel. The helC mutations were present on a plasmid and were tested in the strain ΔhelC. Lanes: 1, ΔhelABC; 2, ΔhelC; 3, pHelC H58G/ΔhelC; 4, pHelC H183G/ΔhelC; 5, pHelC G118A/ΔhelC; 6, ΔhelD; 7, pHelC WT/ΔhelC; 8, SB1003 (Wild type). Light bands above the HelA protein in lanes 5, 7, and 8 are c-type cytochromes. The covalently bound heme of these proteins generates a signal in the ECL assay.
The WWD periplasmic domain of all three classes of proteins is very tryptophan-rich and contains several residues that are conserved in plants and Gram-negative and Gram-positive organisms (Fig. 3). Surprisingly, three mutations of the completely conserved residues within the WWD domain of HelC, W117L, G118A, and D124E resulted in wild-type growth. An helC mutant containing two alterations (G118A/D124E) and ccl1 mutants with defects in the WWD domain (D242E/P243A) also yielded functional proteins by the same type of analysis (Fig. 3). Fig. 4 shows that one of the HelC WWD mutants, G118A, makes a functional complex, as expected.
DISCUSSION
Model for Cytochrome c Biogenesis.
Three systems have developed for the biogenesis of their c-type cytochromes in the three kingdoms of life (33). Organisms and organelles with system I (α and γ proteobacteria and plant and protozoal mitochondria) use the HelABC and Ccl1 proteins. Results from the topological and mutagenic studies reported herein support the hypothesis that this family of proteins is involved in transmembrane heme delivery. The extraordinary conservation of residues observed in the HelBC and Ccl1 proteins at the outer surface of the membrane is also consistent with a delivery and assembly process that takes place at the membrane surface. We propose that reduced heme is exported through the HelABCD transporter to the external WWD domain, where it is liganded by the essential His-1 and His-2 residues (Figs. 1 and 2B). Subsequently, this heme is relayed along the surface of the inner membrane through the Ccl1 protein, retaining histidinyl ligands within the Ccl1 WWD domain. The heme moeity of some heme proteins, such as globins, is rapidly oxidized when a histidinyl ligand has been mutated (34–36). The requirement for heme reduction in cytochrome c biogenesis may necessitate the protection of heme from ambient conditions. Our model suggests that heme is never free in the cytochrome c biogenesis pathway, a conclusion consistent with the results of heme reporter studies in E. coli (37).
We have previously shown that the membrane-tethered thioredox protein Ccl2 specifically reduces apocytochrome c cysteinyl residues (12). Thus, the Ccl2 protein may both present the apocytochrome to the heme and reduce the cysteinyl residues for reaction with heme vinyls. Because the Ccl2 protein is present at more than 20-fold higher levels under oxidative growth conditions, this protein is likely to buffer the apocytochrome c thiols from oxidation (16). The HelX protein, in turn, reduces the Ccl2 protein (12). We now suggest that one explanation for the high conservation of the WWD domain is to retain the heme in the proper orientation for delivery and presentation to the Ccl2/apocytochrome c mixed disulfide at the membrane surface. It is unexpected that changes to conserved residues in the HelC and Ccl1 WWD domain resulted in functional proteins. However, the result that substitutions of absolutely conserved residues in a protein do not lead to protein inactivation has been reported and discussed previously. For example, Kenyon (38) discusses the properties of mutations of highly conserved residues affecting the interconversion of ADP and creatine-phosphate to ATP and creatine by creatine kinase. Likewise, Carreras and Santi (39) present a large number of mutations of highly conserved residues affecting the catalysis of dUMP to dTMP by thymidylate synthase. As with those studies, it is tempting to speculate that the WWD domain affords multiple interactive surfaces to orient the heme molecule for presentation. These multiple surfaces may provide for robustness in function in the HelC and Ccl1 proteins. Alternatively, some conserved residues in the WWD domain might be responsible for structural stability.
Evolutionary Considerations.
Why have two apparently different systems evolved for specific heme delivery in cytochrome c biogenesis (i.e., HelABCD→Ccl1 in some Gram-negative bacteria and plant/protozoal mitochondria and only one WWD-containing protein, CcsA, in Gram-positive bacteria and chloroplasts)? It cannot be ruled out that other proteins may play a role in the CcsA system (e.g., the Ccs1 protein, which is fused to the CcsA protein in Helicobacter pylori; see refs. 33 and 40). If the CcsA protein transports heme across the membrane to present it directly to the apocytochrome c, this could be considered a simplified version of the HelABCD→ Ccl1 heme delivery system. In certain organisms, an energy-dependent (i.e., ATP) transporter may be necessary for rapid cytochrome c synthesis, whereas in Gram-positive bacteria and chloroplasts, simple diffusion through the CcsA protein might be sufficient. The HelABCD transporter may have been recruited for additional functions such as the export of pyoverdine in Pseudomonas (41). Recruitment of ABC transporters for multiple functions is now becoming a more commonly accepted phenomenon (42). Nevertheless, the fundamental roles of the HelABCD and Ccl1 proteins in cytochrome c biogenesis are clearly their primary function. This is consistent with genetic analysis in R. capsulatus whereby strains containing deletions in any of the five genes encoding these proteins have never reverted to cytochrome c proficiency (of greater than 1011 cells tested) (5, 12). The recent discovery by Lang et al. (20) of the helABC and ccl1 genes in one of the most ancestral mitochondrial genomes, that of the protozoan Reclinomonas americana, and the helC and ccl1 genes in the archae (Archaeoglobus) database (ref. 43 and the Institute of Genomic Research genome database) further underscore the pivotal roles of these proteins in the biogenesis process.
Acknowledgments
We thank Karen Gabbert and Asa Flanigan for technical assistance, Diana Beckman for plasmid construction, Dr. John Smith for plasmids, and Josie Clark-Curtiss for M. leprae DNA. We thank Fevzi Daldal, Robert Gennis, Eduardo Groisman, Dewey Holten, and Barbara Kunkel for comments on the manuscript. This research was supported by a grant from the National Institutes of Health (GM47909).
References
- 1.Iwata S, Ostermeier C, Ludwig B, Michel H. Nature (London) 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 2.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 3.Xia D, Yu C A, Kim H, Xia J Z, Kachurin A M, Zhang L, Yu L, Deisenhofer J. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckman D L, Trawick D R, Kranz R G. Genes Dev. 1992;6:268–283. doi: 10.1101/gad.6.2.268. [DOI] [PubMed] [Google Scholar]
- 5.Goldman B S, Beckman D L, Bali A, Monika E M, Gabbert K K, Kranz R G. J Mol Biol. 1997;268:724–738. doi: 10.1006/jmbi.1997.0992. [DOI] [PubMed] [Google Scholar]
- 6.Dumont M E, Ernst J F, Hampsey D M, Sherman F. EMBO J. 1987;6:235–241. doi: 10.1002/j.1460-2075.1987.tb04744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steiner H, Kispal G, Zollner A, Haid A, W, N, Lill R. J Biol Chem. 1996;271:32605–32611. doi: 10.1074/jbc.271.51.32605. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson D W, Neupert W. Proc Natl Acad Sci USA. 1989;86:4340–4344. doi: 10.1073/pnas.86.12.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barker P D, Ferrer J C, Mylrajan M, Leohr T M, Feng R, Konishi Y, Funk W D, MacGillivray R T A, Mauk A G. Proc Natl Acad Sci USA. 1993;90:6542–6546. doi: 10.1073/pnas.90.14.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Page M D, Ferguson S J. Mol Microbiol. 1989;3:653–661. doi: 10.1111/j.1365-2958.1989.tb00213.x. [DOI] [PubMed] [Google Scholar]
- 11.Beckman D L, Kranz R G. Proc Natl Acad Sci USA. 1993;90:2179–2183. doi: 10.1073/pnas.90.6.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monika E M, Goldman B S, Beckman D L, Kranz R G. J Mol Biol. 1997;272:679–692. doi: 10.1006/jmbi.1997.1227. [DOI] [PubMed] [Google Scholar]
- 13.Fabianek R A, Huber-Wunderlich M, Glockshuber R, Künzler P, Hennecke H, Thöny-Meyer L. J Biol Chem. 1997;272:4467–4473. doi: 10.1074/jbc.272.7.4467. [DOI] [PubMed] [Google Scholar]
- 14.Kranz R G. J Bacteriol. 1989;171:456–464. doi: 10.1128/jb.171.1.456-464.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramseier T M, Winteler H V, Hennecke H. J Biol Chem. 1991;266:7793–7803. [PubMed] [Google Scholar]
- 16.Gabbert K K, Goldman B S, Kranz R G. J Bacteriol. 1997;179:5422–5428. doi: 10.1128/jb.179.17.5422-5428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez D H, Bonnard G, Grienenberger J-M. Curr Genet. 1993;24:248–254. doi: 10.1007/BF00351799. [DOI] [PubMed] [Google Scholar]
- 18.Schuster W, Combettes B, Flieger K, Brennicke A. Mol Gen Genet. 1993;239:49–57. doi: 10.1007/BF00281600. [DOI] [PubMed] [Google Scholar]
- 19.Bonnard G, Grienenberger J M. Mol Gen Genet. 1995;246:91–99. doi: 10.1007/BF00290137. [DOI] [PubMed] [Google Scholar]
- 20.Lang B F, Burger G, O’Kelly C J, Cedergren R, Golding G B, Lemieux C, Sankoff D, Turmel M, Gray M W. Nature (London) 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 21.Xie Z, Merchant S. J Biol Chem. 1996;271:4632–4639. doi: 10.1074/jbc.271.9.4632. [DOI] [PubMed] [Google Scholar]
- 22.Sun G, Sharkova E, Chesnut R, Birkey S, Duggan M F, Sorokin A, Pujic P, Ehrlich S D, Hulett F M. J Bacteriol. 1996;178:1374–1385. doi: 10.1128/jb.178.5.1374-1385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perutz M. Annu Rev Physiol. 1990;52:1–25. doi: 10.1146/annurev.ph.52.030190.000245. [DOI] [PubMed] [Google Scholar]
- 24.Morgan W T, Muster P, Tatum F, Kao S M, Alam J, Smith A. J Biol Chem. 1993;268:6256–62. [PubMed] [Google Scholar]
- 25.Barnes W. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiedeman A A, Smith J M. Nucleic Acids Res. 1988;16:3587. doi: 10.1093/nar/16.8.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manoil C, Beckwith J. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 29.Lang S E, Jenney F E, Jr, Daldal F. J Bacteriol. 1996;178:5279–5290. doi: 10.1128/jb.178.17.5279-5290.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikaido H, Saier M. Science. 1992;258:936–942. doi: 10.1126/science.1279804. [DOI] [PubMed] [Google Scholar]
- 31.Froshauer S, Green G N, Boyd D, McGovern K, Beckwith J. Mol Microbiol. 1988;1:101–106. doi: 10.1016/0022-2836(88)90539-6. [DOI] [PubMed] [Google Scholar]
- 32.Handa H, Bonnard G, Grienenberger J M. Mol Gen Genet. 1996;252:292–302. doi: 10.1007/BF02173775. [DOI] [PubMed] [Google Scholar]
- 33.Goldman B S, Kranz R G. Mol Microbiol. 1998;27:871–873. doi: 10.1046/j.1365-2958.1998.00708.x. [DOI] [PubMed] [Google Scholar]
- 34.Nagai K, Luisi B, Shih D, Miyazaki G, Imai K, Poyart C, De Young A, Kwiatkowsky L, Noble R W, Lin S H. Nature (London) 1987;329:858–860. doi: 10.1038/329858a0. [DOI] [PubMed] [Google Scholar]
- 35.Olson J S, Mathews A J, Rohlfs R J, Springer B A, Egeberg K D, Sligar S G, Tame J, Renaud J P, Nagai K. Nature (London) 1988;336:265–266. doi: 10.1038/336265a0. [DOI] [PubMed] [Google Scholar]
- 36.Shelver D, Kerby R L, He Y, Roberts G P. Proc Natl Acad Sci USA. 1997;94:11216–11220. doi: 10.1073/pnas.94.21.11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldman B S, Gabbert K K, Kranz R G. J Bacteriol. 1996;178:6338–6347. doi: 10.1128/jb.178.21.6338-6347.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kenyon G L. Nature (London) 1996;381:281–282. doi: 10.1038/381281a0. [DOI] [PubMed] [Google Scholar]
- 39.Carreras C W, Santi D V. Annu Rev Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 40.Kranz, R. G., Lill, R., Goldman, B. S., Bonnard, G. & Merchant, S. (1998) Mol. Microbiol., in press. [DOI] [PubMed]
- 41.Gaballa A, Koedam N, Cornelis P. Mol Microbiol. 1996;21:777–785. doi: 10.1046/j.1365-2958.1996.391399.x. [DOI] [PubMed] [Google Scholar]
- 42.Higgins C F. Cell. 1995;82:693–696. doi: 10.1016/0092-8674(95)90465-4. [DOI] [PubMed] [Google Scholar]
- 43.Klenk H-P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. Nature (London) 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]