Abstract
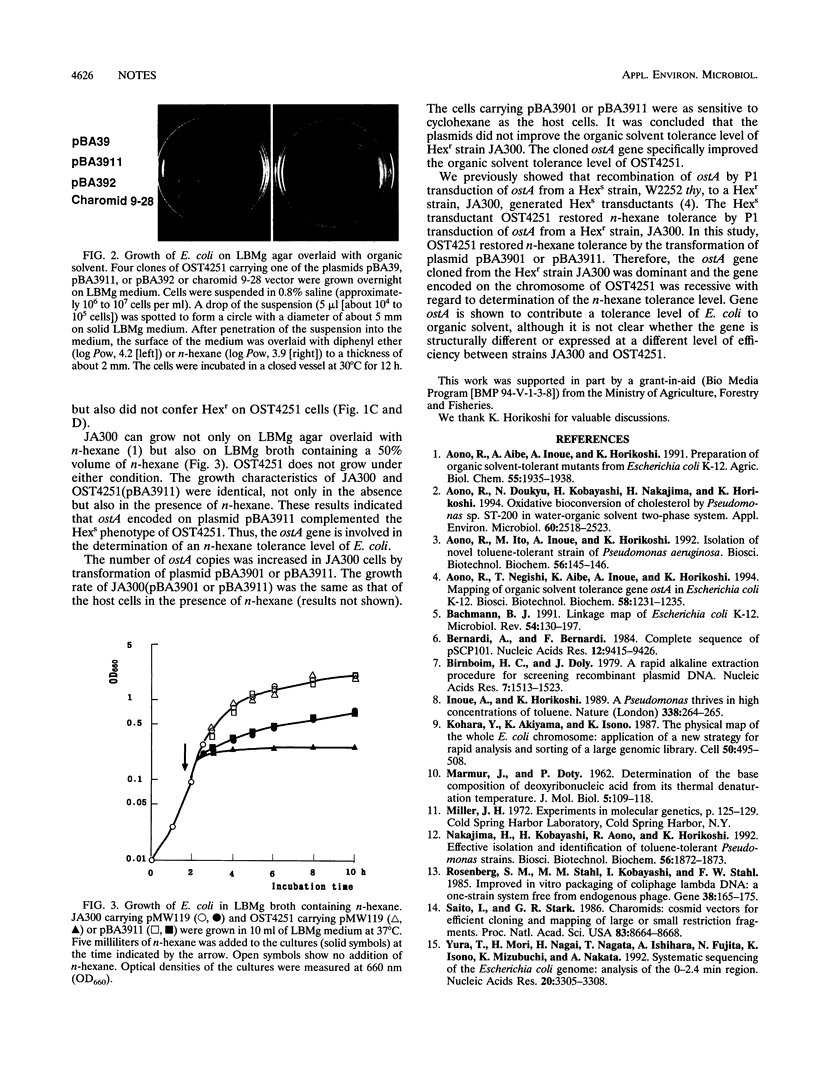
A variety of genes are involved in determining the level of organic solvent tolerance of Escherichia coli K-12. Gene ostA is one of the genes contributing to the level of organic solvent tolerance. This gene was cloned from an n-hexane-tolerant strain of E. coli, JA300. A JA300-based n-hexane-sensitive strain, OST4251, was converted to the n-hexane-tolerant phenotype by transformation with DNA containing the ostA gene derived from JA300. Thus, the cloned ostA gene complemented the n-hexane-sensitive phenotype of OST4251.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aono R., Doukyu N., Kobayashi H., Nakajima H., Horikoshi K. Oxidative Bioconversion of Cholesterol by Pseudomonas sp. Strain ST-200 in a Water-Organic Solvent Two-Phase System. Appl Environ Microbiol. 1994 Jul;60(7):2518–2523. doi: 10.1128/aem.60.7.2518-2523.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono R., Negishi T., Aibe K., Inoue A., Horikoshi K. Mapping of organic solvent tolerance gene ostA in Escherichia coli K-12. Biosci Biotechnol Biochem. 1994 Jul;58(7):1231–1235. doi: 10.1271/bbb.58.1231. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi A., Bernardi F. Complete sequence of pSC101. Nucleic Acids Res. 1984 Dec 21;12(24):9415–9426. doi: 10.1093/nar/12.24.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg S. M., Stahl M. M., Kobayashi I., Stahl F. W. Improved in vitro packaging of coliphage lambda DNA: a one-strain system free from endogenous phage. Gene. 1985;38(1-3):165–175. doi: 10.1016/0378-1119(85)90215-x. [DOI] [PubMed] [Google Scholar]
- Saito I., Stark G. R. Charomids: cosmid vectors for efficient cloning and mapping of large or small restriction fragments. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8664–8668. doi: 10.1073/pnas.83.22.8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T., Mori H., Nagai H., Nagata T., Ishihama A., Fujita N., Isono K., Mizobuchi K., Nakata A. Systematic sequencing of the Escherichia coli genome: analysis of the 0-2.4 min region. Nucleic Acids Res. 1992 Jul 11;20(13):3305–3308. doi: 10.1093/nar/20.13.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]




