Abstract
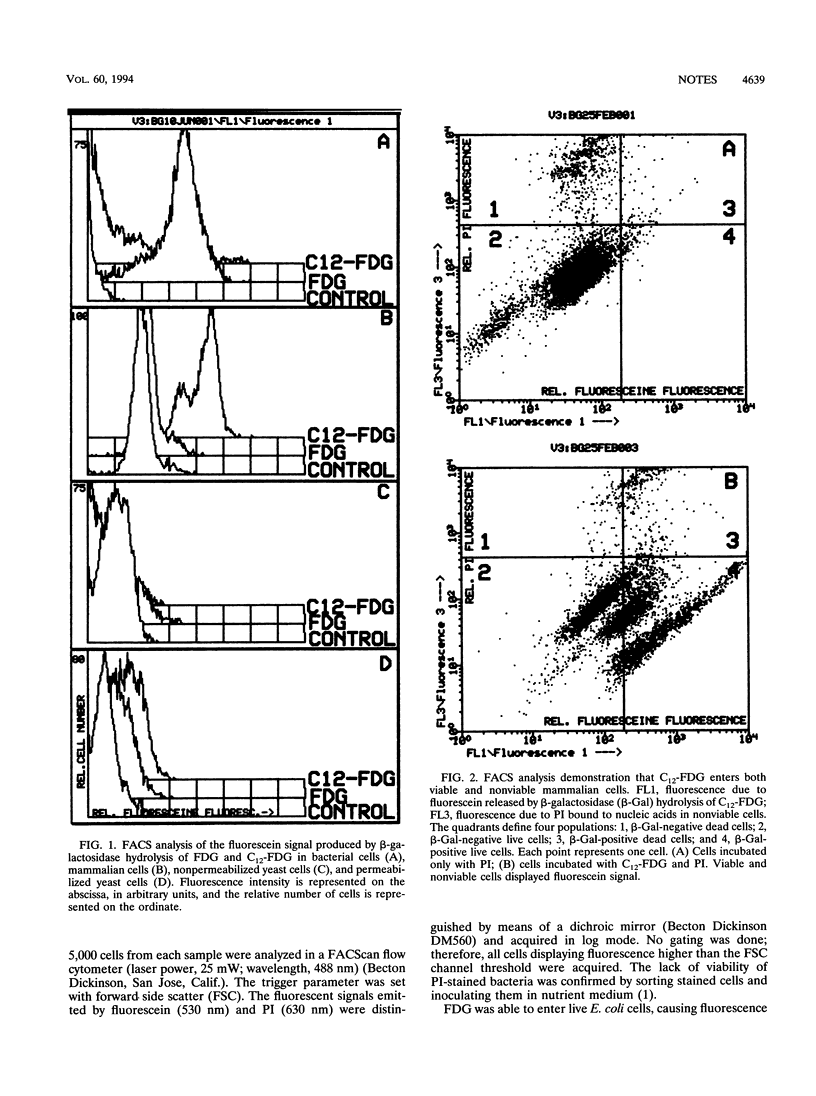
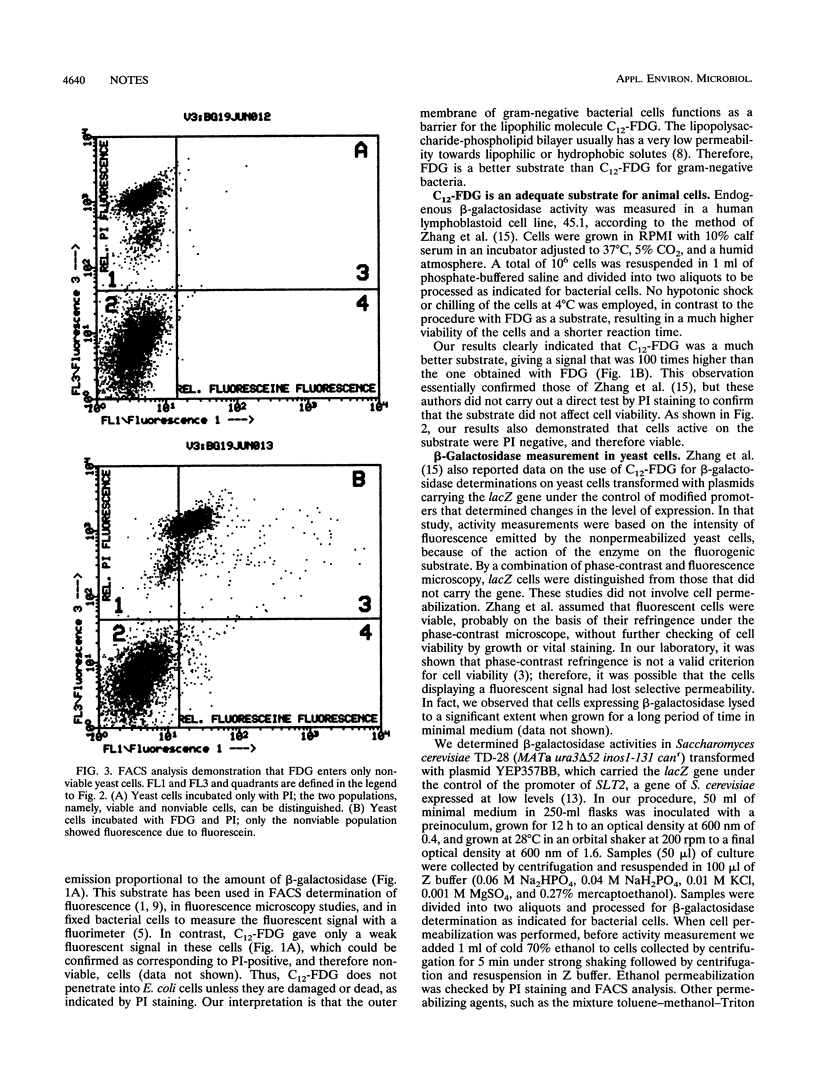
Fluorescein-di-beta-D-galactopyranoside (FDG) was found to be a useful substrate for beta-galactosidase detection by flow cytometry in gram-negative bacteria, since it entered viable cells and gave a fluorescence emission proportional to the enzymatic activity. C12-FDG, a more lipophilic derivative, gave a very poor signal because of the lack of penetration. On the contrary, C12-FDG was more sensitive than FDG for beta-galactosidase activity determinations in animal cells. In contrast to previous reports, C12-FDG did not enter viable yeast cells, so that the use of the substrate required cell permeabilization. Without this treatment, C12-FDG penetrates only nonviable yeast cells that may occur in populations expressing beta-galactosidase.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alvarez A. M., Ibáez M., Rotger R. Beta-galactosidase activity in bacteria measured by flow cytometry. Biotechniques. 1993 Dec;15(6):974–976. [PubMed] [Google Scholar]
- Fiering S. N., Roederer M., Nolan G. P., Micklem D. R., Parks D. R., Herzenberg L. A. Improved FACS-Gal: flow cytometric analysis and sorting of viable eukaryotic cells expressing reporter gene constructs. Cytometry. 1991;12(4):291–301. doi: 10.1002/cyto.990120402. [DOI] [PubMed] [Google Scholar]
- Ibáez M., Rotger R. Characterization of a small cryptic plasmid from Salmonella enteritidis that affects the growth of Escherichia coli. FEMS Microbiol Lett. 1993 May 15;109(2-3):225–229. doi: 10.1111/j.1574-6968.1993.tb06172.x. [DOI] [PubMed] [Google Scholar]
- Nir R., Yisraeli Y., Lamed R., Sahar E. Flow cytometry sorting of viable bacteria and yeasts according to beta-galactosidase activity. Appl Environ Microbiol. 1990 Dec;56(12):3861–3866. doi: 10.1128/aem.56.12.3861-3866.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan G. P., Fiering S., Nicolas J. F., Herzenberg L. A. Fluorescence-activated cell analysis and sorting of viable mammalian cells based on beta-D-galactosidase activity after transduction of Escherichia coli lacZ. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2603–2607. doi: 10.1073/pnas.85.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M., Botstein D. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- Srienc F., Campbell J. L., Bailey J. E. Flow cytometry analysis of recombinant Saccharomyces cerevisiae populations. Cytometry. 1986 Mar;7(2):132–141. doi: 10.1002/cyto.990070203. [DOI] [PubMed] [Google Scholar]
- Torres L., Martín H., García-Saez M. I., Arroyo J., Molina M., Sánchez M., Nombela C. A protein kinase gene complements the lytic phenotype of Saccharomyces cerevisiae lyt2 mutants. Mol Microbiol. 1991 Nov;5(11):2845–2854. doi: 10.1111/j.1365-2958.1991.tb01993.x. [DOI] [PubMed] [Google Scholar]
- Wittrup K. D., Bailey J. E. A single-cell assay of beta-galactosidase activity in Saccharomyces cerevisiae. Cytometry. 1988 Jul;9(4):394–404. doi: 10.1002/cyto.990090418. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Z., Naleway J. J., Larison K. D., Huang Z. J., Haugland R. P. Detecting lacZ gene expression in living cells with new lipophilic, fluorogenic beta-galactosidase substrates. FASEB J. 1991 Dec;5(15):3108–3113. doi: 10.1096/fasebj.5.15.1720751. [DOI] [PubMed] [Google Scholar]
- de la Fuente J. M., Alvarez A., Nombela C., Sanchez M. Flow cytometric analysis of Saccharomyces cerevisiae autolytic mutants and protoplasts. Yeast. 1992 Jan;8(1):39–45. doi: 10.1002/yea.320080104. [DOI] [PubMed] [Google Scholar]



