Summary
Glycoprotein K (gK) is a virion envelope component of herpes simplex virus types 1 (HSV-1) and 2 (HSV-2), which plays an important role in virion morphogenesis and egress. We previously demonstrated that immunization of mice with gK, but not with any of the ten other HSV-1 glycoproteins, resulted in exacerbation of corneal scarring and herpetic dermatitis following ocular HSV-1 infection. However, little is known about the gK epitope(s) that is (are) involved in T cell activities in vitro or in vivo. Thus, epitope mapping of gK was performed using a panel of 15-mer peptides with five-amino-acid overlaps spanning the full-length gK, and four expressed gK recombinant proteins representing different regions of gK. Epitope mapping within the gK polypeptide defined the amino acid sequence STVVLITAYGLVLVW as the predominant CD4+ and CD8+ T cell stimulatory region both in vitro and in vivo. IFN-γ expression by CD4+ T cells was CD8+ T cells-dependent. This immunodominant epitope is located within the signal sequence of the gK polypeptide and is highly conserved in HSV-1 and HSV-2 strains. Using prediction algorithms, the peptide is predicted to bind to numerous MHC class I and class II molecules.
Keywords: CTL, IFN-γ, Proliferation, CFSE, Peptide, T cells, MHC
Introduction
Glycoprotein K (gK) is one of the 11 known HSV-1 glycoproteins (Ghiasi et al., 1994b; Hutchinson et al., 1992; McGeoch et al., 1988). gK is encoded by the UL53 open reading frame and is a highly hydrophobic 338-amino-acid protein with a predicted molecular mass of 37-kDa (McGeoch et al., 1988). Both gK from HSV-1 and HSV-2 are 338 amino acids long with an approximately 84% amino acid homology (Dolan et al., 1998; McGeoch et al., 1991; McGeoch et al., 1988). gK has a cleavable 30-amino-acid NH2-terminal signal sequence and it is glycosylated at two potential sites for N glycosylation on amino acids 48 and 58 (Debroy et al., 1985; McGeoch et al., 1988; Ramaswamy and Holland, 1992). In HSV-1 infected cells, gK is expressed as a 39 kDa high-mannose precursor polypeptide, designated precursor gK (pgK), which is further glycosylated to produce a 41 kDa protein mature glycoprotein (Hutchinson et al., 1992). Furthermore, when gK is expressed by a recombinant baculovirus, four gK-related recombinant baculovirus-expressed polypeptides of 29-, 35-, 38-, and 40-kDa were detected (Ghiasi et al., 1994b). The 35-, 38-, and 40 kDa species were susceptible to tunicamycin inhibition revealing that they were N-linked glycosylated. The 38- and 40-kDa proteins corresponded to pgK and mature gK, respectively, while the 29-kDa protein represented the cleaved, unglycosylated peptide and the 35-kDa protein represented the cleaved and partially glycosylated peptide. gK translated in vitro had a molecular mass of 36-kDa with three possible membrane-spanning regions (Mo and Holland, 1997; Ramaswamy and Holland, 1992). In line with this study, recently we have constructed a baculovirus recombinant gK that had the 163 amino acids from its carboxy-terminal end removed. However, despite the removal of two of the three possible membrane-spanning regions this peptide of 175 amino acids long was not secreted (Ghiasi, et al, unpublished).
gK is thought to be an important determinant of virus-induced cell fusion, since single amino acid changes within gK cause extensive virus-induced cell fusion (Bond and Person, 1984; Debroy et al., 1985; Little and Schaffer, 1981; Pogue-Geile and Spear, 1987). Furthermore, gK is an important determinant of cytoplasmic virion envelopment, since viruses lacking gK fail to efficiently acquire a cytoplasmic envelope resulting in a drastic defect in virion egress and spread (Foster and Kousoulas, 1999; Hutchinson and Johnson, 1995; Hutchinson et al., 1995). Similar to HSV-1 gK, pseudorabies and varicella-zoster virus gKs also play an important role in virion morphogenesis and egress (Dietz et al., 2000; Klupp et al., 1998; Mo et al., 1999).
Using anti-gK peptide antibodies it was shown that gK is expressed within the endoplasmic reticulum (ER) and nuclear membranes of infected cells (Hutchinson et al., 1995), while recently, it was shown that gK is expressed on infected cell surfaces (Foster et al., 2003). Also, intracellular transport, Trans-Golgi Network (TGN) localization and cell-surface expression of gK required the presence of the UL20 protein indicating that gK and UL20 protein functionally interact (Foster et al., 2004).
We previously demonstrated that immunization of mice with gK, but not with any of the ten other HSV-1 glycoproteins, resulted in exacerbation of corneal scarring (CS) and dermatitis following ocular HSV-1 infection (Ghiasi et al., 1995; Ghiasi et al., 1994a). The exacerbation of CS and dermatitis in gK-immunized mice was due to the presence of CD8+ T cells in the cornea of ocularly infected mice (Osorio et al., 2004). The region of gK that contributes to exacerbation of CS and dermatitis and induction of CD8+ T cell response is not known. To identify the region of gK that is involved in this process, we have synthesized a panel of 33 peptides expanding the 338 amino acids of the gK polypeptide. From these studies, we have identified a single epitope within the amino-terminal region of gK. This peptide is involved in T cell proliferation and IFN-γ expression in vitro and CTL activity in vivo. These responses are CD8-dependent.
Materials and Methods
Virus and cells
Triple plaque-purified HSV-1 strain McKrae was grown in RS (rabbit skin) cell monolayers in minimal essential media (MEM) containing 10% fetal calf serum (FCS) as was previously described (Ghiasi et al., 1994b). Spleen cells were grown in RPMI1640 medium supplemented with 10% FCS.
Expression of HSV-1 glycoprotein K (gK) in insect cells
Construction and characterization of a full-length gK (defined in this study as gK338) was described previously (Ghiasi et al., 1994b). An additional recombinant baculovirus was constructed that expressed the amino terminal 175 amino acids of gK. This construct was named gK175. Sf9 insect cells were infected with 10 PFU/cell of gK338, or gk175 for 72 hr, as was described previously (Ghiasi et al., 1994b). gK338 and gK175 were affinity-purified from cell lysates of Sf9 infected-cells using a rabbit polyclonal antibody against a 20-mer (LIAGRVVPFQVP PDAMNRRI, UCLA Peptide Synthesis Core Facility) peptide of gK deduced amino acids (McGeoch et al., 1988) and were dissolved in DMSO at a concentration of 10 μg/μl for splenocytes stimulation. Both gK338 and gK175 were found to be cell-associated.
Expression of HSV-1 gK in E. coli
The cDNA for full-length gK (Ghiasi et al., 1994b) was inserted into several E. coli expression vectors, but due to severe toxicity associated with full-length gK, we used different fragments of gK for expression in E. coli. Briefly, the gK ORF was digested into three fragments called D1, D2, and D3. D1 consists of amino acids 1 to 175 (similar fragment that was used for baculovirus-expressed gK175). D2 is 88 amino acids long, consists of amino acids 148 to 237, and encodes a protein of approximately 10.1-kDa. D3 is 71 amino acids long, consists of amino acids 229 to 300, and it encodes a protein of approximately 7.6-kDa. D1 was toxic in E. coli; however, D2 and D3 were not. The purified D2 and D3 polypeptides were dissolved in DMSO at a concentration of 10 μg/μl.
Mice
Inbred BALB/c (female, 6 weeks old) mice were obtained from The Jackson Laboratory.
Immunization
BALB/c mice were vaccinated three times at two week intervals subcutaneously (SC) with baculovirus recombinant expressing gK338 infected cell lysates (Ghiasi et al., 1994b). Each vaccination consisted of extracts from 1 × 106 cells, which contained 5 μg of gK, based on the intensity of the expressed gK band on stained gels. The mock vaccine consisted of extract from the same number of cells infected with wild-type baculovirus. The first SC injection was in Freund's complete adjuvant. Subsequent vaccinations used Freund's incomplete adjuvant. As positive controls, some mice were immunized 3X with 1×106 PFU of HSV-1 strain KOS IP.
Peptide synthesis
A panel of 33 overlapping HSV-1 gK peptides (15-mers with five-amino-acid overlaps) were synthesized by Mimotopes (San Diego, CA) and are: #1) MLAVRSLQHLSTVVL; #2) STVVLITAYGLVLVW; #3) LVLVWYTVFGASPLH; #4) ASPLHRCIYAVRPTG; #5) VRPTGTNNDTALVWM; #6) ALVWMKMNQTLLFLG; #7) LLFLGAPTHPPNGGW; #8) PNGGWRNHAHICYAN; #9) ICYANLIAGRVVPFQ; #10) VVPFQVPPDAMNRRI; #11) MNRRIMNVHEAVNCL; #12) AVNCLETLWYTRVRL; #13) TRVRLVVVGWFLYLA; #14) FLYLAFVALHQRRCM; #15) QRRCMFGVVSPAHKM; #16) PAHKMVAPATYLLNY; #17) YLLNYAGRIVSSVFL; #18) SSVFLQYPYTKITRL; #19) KITRLLCELSVQRQN; #20) VQRQNLVQLFETDPV; #21) ETDPVTFLYHRPAIG; #22) RPAIGVIVGCELMLR; #23) ELMLRFVAVGLIVGT; #24) LIVGTAFISRGACAI; #25) GACAITYPLFLTITT; #26) LTITTWCFVSTIGLT; #27) TIGLTELYCILRRGP; #28) LRRGPAPKNADKAAA; #29) DKAAAPGRSKGLSGV; #30) GLSGVCGRCCSIILS; (#31) SIILSGIAVRLCYIA; #32) LCYIAVVAGVVLVAL; and #33) VLVALHYEQEIQRRL. These 33 peptides cover the entire gK protein sequence. Purity of original peptides synthesized was at least 70%. All peptides were dissolved in DMSO at a concentration of 1 μg/μl and stored at −20°C.
Lymphocyte proliferation response
Spleens from immunized mice were removed, and single cell suspensions were prepared. Isolated lymphocytes were resuspended at 1 × 105 cells/100 μl in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum and stimulated in vitro with each peptide or 10 PFU/cell of UV-inactivated McKrae for 48 hr. On day 2, 1 μl of UdBr was added to 1 × 105 lymphocytes. Incorporation of UdBr was determined 24 hr later, as described by the manufacturer (EMD Biosciences, San Diego, CA). Controls included unstimulated lymphocytes from immunized mice and lymphocytes from naive mice.
IFN-γ assay
Single cell suspensions of lymphocytes from gK-immunized or control-immunized mice were resuspended at 2 × 106 cells/500 μl in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum. Cells were cultured in triplicate in the presence or absence of 1, 5, 10, 50, or 100 μg of each peptide for three days at 37°C in 5% CO2. As positive controls, spleens were stimulated with 10 PFU/cell of UV-inactivated HSV-1 strain McKrae or concanavalin A (ConA). ConA was used at a concentration of 1 or 0.1 μg/2 × 106 cells. Mock-immunized (mouse spleens?) were used as negative controls and were similarly stimulated with individual synthetic peptides. Culture supernatants were collected three days after incubation, and the concentration of IFN-γ was determined by enzyme-linked immunosorbent assay (ELISA) (Becton Dickinson, San Diego, CA).
Determination of IFN-γ producing CD4+ and CD8+ T cells
Single cell suspensions of spleen cells from individual mice were prepared as above and described previously (Ghiasi et al., 1994a). Isolated spleen cells at a concentration of 2 × 106 cells/500 μl of cell culture media were stimulated with 10 μg of each synthetic peptide for 72 hr. Control spleens were stimulated with UV-inactivated HSV-1, ConA (1 μg/500 μl), or no stimulation. Four hours before collecting stimulated spleen cells, cells were pulsed with 2.5 μg/ml of brefeldin A (BFA) for 4 hr. Spleen cells stimulated with each peptide were harvested and divided into two groups, half of each sample was immunostained with PE-conjugated anti-L3T4 (anti-CD4) mAb and the other half was stained with PE-conjugated anti-Lyt-2 (anti-CD8) mAb (BD PharMingen, San Diego, CA), according to the manufacturer's recommendations. For IFN-γ intracellular staining, cells were washed, fixed, and permeabilized with Cytofix/Cytoperm solution (BD Biosciences, San Diego, CA) for 30 min at 4°C. The cells were then washed in Cytofix/Cytoperm solution and incubated with FITC-labeled IFN-γ mAb (BD PharMingen) for 15 min at 4°C. After two washes with permeabilizing buffer, the cells were analyzed via FACS. Double-color flow cytometric analyses of cells were performed using a FACScan (Beckton Dickinson, PA). Controls included cells incubated with an irrelevant antibody. Experiments were repeated twice.
Isolation of individual T cell populations
To determine if stimulation of T cells by peptide #2 is associated with CD4+ T cells alone or CD8+ T cells alone, the single cell suspensions of lymphocytes from gK or control-immunized mice were prepared as described above. For isolation of CD4+ and CD8+ T cells, lymphocytes from two mice were combined and divided into two parts. One part was run on a CD4+ T cell enrichment column, while the other half was run thru a CD8+ T cell enrichment column (R&D System, MN). To completely eliminate the possible contamination of CD4+ cells in the CD8+ group and CD8+ cells from the CD4+ group, the isolated cells were incubated with 100 μg of anti-CD4+ or anti-CD8+ mAb for 30 min on ice. Following this treatment, each cell population was run through their respective column again. This procedure rendered each isolated T cell population free of contamination from the other population and >97% purified as determined by FACS. The number of column-purified CD4+ and CD8+ T cells was determined, and 1 × 106 cells in 200 μl of cell culture media were stimulated with 10 μg of each synthetic peptide for 72 hr. Expression of IFN-γ by each T cell subpopulation was determined by ELISA as described above.
In vivo measurement of cytotoxicity
Single cell suspensions of spleen cells from naive BALB/c mice were prepared as above and used as target cells to detect in vivo cytotoxic activity. Spleen cells from each mouse were divided into two populations. One population was pulsed with 10 μg of peptide #1 or #2, incubated at 37°C for 60 min, and labeled with a high concentration of 5-(6)-carboxy-fluorescein succinimidyl ester (CFSE) (2.5 μM) (CFSEhigh cells). The second control target population was pulsed with 10 μg of nonspecific control VSV peptide (YTDIEMNRLGK) (Sigma-Aldrich) and was labeled with a low concentration of CFSE (0.25 μM) (CFSElow cells). As an unprimed control, splenocytes from naive mice were divided into two populations and stained without any priming. Following CFSE labeling, the two populations of high and low CFSE-labeled lymphocytes were combined, and the equivalent of one mouse's worth of lymphocytes in 100 μl of PBS was then injected intravenously (i.v.) into each recipient mouse that had been previously immunized with gK, KOS, or mock-immunized as described above. Recipient mice were sacrificed four hrs after adoptive transfer of lymphocytes, and their spleens were isolated. Single cell suspensions of lymphocytes were analyzed by FACS and each population was detected by its differential CFSE fluorescence intensity. Specific lysis for each peptide was measured as follows: ratio = (CFSElow%/CFSEhigh%). Percentage specific lysis = [1 − (ratio unprimed/ratio primed)] × 100 (Kassim et al., 2006). To rule out the possible involvement of the other 31 gK peptides in the in vivo cytotoxicity, we grouped the remaining 31 peptides into four groups of seven or eight peptides each, and we performed additional experiments as described above for peptides #1 and #2. The four groups were as follows: Group 1 (peptides #3 to 9), group 2 (peptides # 10 to 17), group 3 (peptides # 18 to 25), and group 4 (peptides # 26 to 33). Similar to peptide #1, none of the gK-immunized mice that received T cells from Group 1, 2, 3, or 4 showed any significant lysis in vivo, thus as a negative control, we only shown the data for peptide #1 in Fig. 5.
Prediction of MHC I and MHC II peptide-binding sites
In silico analysis were used to predict putative MHC class I or MHC class II restricted T cell epitopes. Initial evaluation of the gK complete amino acid sequence was aimed to identify promiscuous MHC binding peptides using virtual matrices designed for several HLA alleles (Mustafa and Shaban, 2006; Singh and Raghava, 2001). To maximize prediction accuracy for MHC class I molecules, we used the following computational systems: SYFPEITHI (Rammensee et al., 1999), MHCPRED (Guan et al., 2003), RANKPEP (Reche et al., 2004) and the Bioinformatics and Molecular Analysis Section (BIMAS) HLA Peptide Binding Predictions (Parker et al., 1994).
Statistical analysis
Protective parameters were analyzed by the Student's t test and Fisher's exact test using Instat (GraphPad, San Diego, CA). Results were considered statistically significant when the “P” value was <0.05.
Results
Lymphocyte proliferation
Induction of lymphocyte proliferation response by each of the 33 synthetic peptides was determined as described in Materials and Methods, using spleen cells from gK- or mock-immunized mice. Following stimulation with each individual peptide, only gK lymphocytes that were stimulated with peptide #2 showed significant incorporation of UdBr compared with the other 32 peptides (Table 1). These differences between peptide #2 and the other 32 peptides were statistically significant (P<0.0001, student t-test). None of the lymphocytes from mock-immunized mice showed any significant increase in incorporation of UdBr following stimulation with any of the peptides including #2 peptide (Table 1). As expected, ConA stimulated lymphocytes from both groups, while UV-inactivated HSV-1 stimulated the gK-immunized but not the mock-immunized group (Table 1). Thus, these results suggest that among the 33 synthetic peptides used to stimulate gK lymphocytes, only peptide #2 had proliferative activity in vitro.
Table 1. Proliferation of gK-immunized T cells following stimulation with gK synthetic peptides.a.
| Immunized | |||
|---|---|---|---|
| Stimulator | gK | Mock | p-value |
| Peptide #2 | 1.06±0.04 | 0.1±0.02 | <0.0001 |
| Remaining 32 peptidesb | 0.20±0.06 | 0.12±0.07 | 0.40 |
| Con A | 1.2±0.32 | 1.0±0.37 | 0.69 |
| HSV-1 | 1.1±0.13 | 0.04±0.01 | <0.0001 |
| VSV | 0.10±0.04 | 0.06±0.02 | 0.38 |
| No stimulation | 0.08±0.03 | 0.11±0.04 | 0.56 |
Splenocytes were primed in vitro for 48 hr, and cells were labeled with UdBr for 24 hr as described in Materials and Methods. Each point represents mean ± SEM for nine replicates from three mice. Proliferations are shown as UdBr absorbance at 450 nm.
Following stimulation, none of the remaining 32 peptides stimulated the gK- or mock-immunized lymphocytes, and proliferation following stimulation with these 32 peptides is shown as one group.
IFN-γ production following stimulation with synthetic peptides
To determine if any of the 33 synthetic peptides stimulated IFN-γ production in vitro, BALB/c mice were immunized with baculovirus-expressed gK or mock-immunized with wt baculovirus as described in Materials and Methods. Splenocytes were isolated from the spleens of gK- or mock-immunized mice three weeks after the third immunization, and 2 × 106 splenocytes per well were stimulated with 10 μg of each of the 33 synthetic peptides. As controls, splenocytes were stimulated with 10 PFU/cell of UV-inactivated McKrae, 1 μg of ConA, 10 μg of irrelevant VSV peptide, or nothing. Seventy-two hours after in vitro stimulation, cell culture supernatants were harvested for analysis of IFN-γ production by ELISA as described in Materials and Methods.
Overall, stimulation of splenocytes from gK-immunized mice with peptide #2 led to the production of IFN-γ in vitro (Table 2). Splenocytes from mock-immunized control mice did not produce any significant level of IFN-γ (Table 2). This difference between IFN-γ productions by splenocytes from gK-immunized mice compared with mock-immunized mice following stimulation with peptide #2 was statistically significant (p < 0.0001). In contrast to peptide #2, spleen cells from gK-immunized mice did not appear to produce any IFN-γ following stimulation with the remaining 32 peptides (Table 2). Splenocytes from both gK and mock groups produced similar levels of IFN-γ following stimulation with ConA (Table 2; p = 0.35). Similar to peptide #2, stimulation of gK splenocytes with UV-inactivated virus produced a significantly higher level of IFN-γ production than splenocytes from mock-immunized mice (Table 2; p = 0.001). However, the level of IFN-γ production in splenocytes from gK-immunized mice was significantly higher following stimulation with peptide #2 than following stimulation with UV-inactivated virus (229±11 vs. 41±8) (Table 2; p = <0.0001). These differences are probably due to a higher concentration of gK active domain using the synthetic peptide than with using the whole virus. Finally, the irrelevant VSV peptide (Table 2; p = 0.28) or no stimulation (Table 2; p = 0.36) did not induce any significant level of IFN-γ in the gK group compared with the mock group. Thus, the kinetics of IFN-γ production in splenocytes of gK-immunized mice following stimulation with peptide #2 and UV inactivated virus appeared to be different from that of the other 32 gK peptides.
Table 2. IFN-γ expression of gK-immunized lymphocytes following stimulation with gK synthetic peptides.a.
| Immunized | |||
|---|---|---|---|
| Stimulator | gK | Mock | p-value |
| Peptide # 2 | 229±11 | 10±4 | <0.0001 |
| Remaining 32 peptidesb | 4±1 | 9±4 | 0.39 |
| Con A | 653±192 | 451±87 | 0.35 |
| HSV-1 | 41±8 | 9±2 | 0.001 |
| VSV-peptide | 11±3 | 7±2 | 0.28 |
| No stimulation | 1±0.3 | 0.4±0.1 | 0.36 |
Splenocytes were primed in vitro for 72 hr, and IFN-γ was measured by ELISA as described in Materials and Methods. Each point represents mean ± SEM for nine replicates from three mice.
Following stimulation, all of the remaining 32 peptides produced less than 5 picogram/ml of IFN-γ following stimulation of gK- or mock-immunized lymphocytes. IFN-γ produced by the 32 peptides is presented as one group.
Our epitope mapping study suggests that peptide #2, which corresponds to amino acids 11 to 25 of gK, has proliferative and IFN-γ activities following stimulation of splenocytes from gK-immunized mice in vitro. To rule out the possible involvement of the carboxy-terminal region of gK in T cell activities in vitro, we used a truncated gK protein of 175 aa that contains peptide #2 and lacks 179 aa from the carboxy-terminal end. In addition, we used D2 (Domain 2) and D3 (Domain 3) corresponding to aa 148-237 and aa 229-300, respectively. Full-length gK (gK338) was used as a positive control. Lymphocytes from gK- and mock-immunized mice were stimulated with gK338, gK175, D2, or D3 as described in Materials and Methods.
As expected, both gK338 and gK175 stimulated gK lymphocytes and produced IFN-γ (Fig. 1). The amount of IFN-γ produced by gK338 vs. gK175 was not statistically significant (p = 0.6). In contrast, gK splenocytes following stimulation with neither D2 nor D3 produced any IFN-γ (Fig. 1). The differences between IFN-γ production following stimulation with gK338 and gK175 compared with IFN-γ production following stimulation with D2 or D3 were statistically significant (Fig. 1, p < 0.003). These results suggested that the gK region that is involved in IFN-γ production is located within the first 175 amino acids of gK. In addition, by using these larger fragments of gK we explored the contribution of secondary structures in non-linear epitopes. The synthetic peptides fully mimic the activity of intact molecule, suggesting that secondary structure may not be involved in the generation of epitopes.
Fig. 1. IFN-γ production by splenocytes of gK-immunized mice following stimulation with different fragments of expressed gK.
Splenocytes from gK-immunized mice were stimulated with purified gK338, gK175, D2, or D3 in vitro for 72 hr as described in Materials and Methods. The concentrations of IFN-γ in the supernatants were measured by ELISA. Each point represents mean ± SEM for nine replicates from three mice.
Detection of IFN-γ producing CD4+ and CD8+ T cells following stimulation of gK splenocytes with peptide #2
Mice were immunized with gK, KOS, or gD, or they were mock-immunized. Three weeks after the third vaccination or mock-vaccination, splenocytes isolated from immunized or mock-immunized BALB/c mice were stimulated with 10 μg of each of the 33 peptides, VSV peptide, UV-inactivated HSV-1 strain McKrae, ConA, or nothing as described in Materials and Methods. Four hours before harvesting the cells, BFA was added to each well, and cells in suspension were immunostained with PE-conjugated anti-L3T4 (anti-CD4) mAb or PE-conjugated anti-Lyt-2 (anti-CD8) mAb as described in Materials and Methods. Following staining for CD4 or CD8, the cells were stained intracellularly with FITC-conjugated anti-IFN-γ mAb. Double-color flow cytometric analyses of total spleen cells were performed using a FACScan (Beckton Dickinson, PA). Controls included cells incubated with an irrelevant antibody.
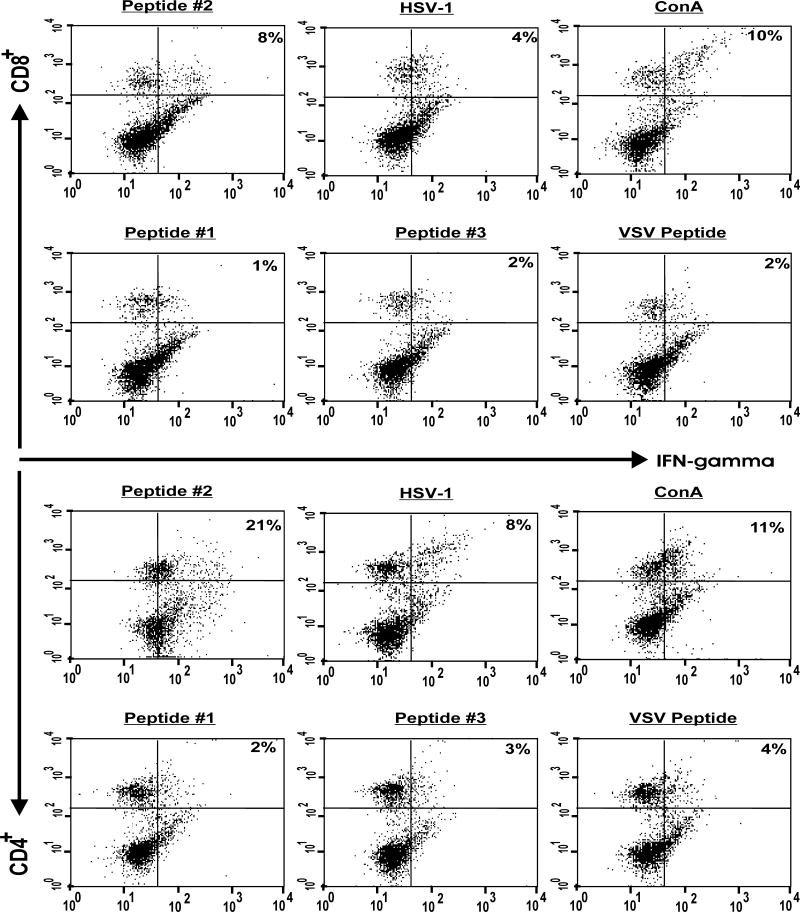
Splenocytes from gK-immunized mice that were stimulated with peptide #2 showed significantly higher percentages of CD4+IFN-γ+ (21%) and CD8+IFN-γ+ (8%) than splenocytes stimulated with peptides #1 and #3 (Fig. 2). Similarly, following stimulation of gK splenocytes with HSV-1, the levels of CD4+IFN-γ+ and CD8+IFN-γ+ cells were increased but the percentages of CD4+IFN-γ+ and CD8+IFN-γ+ were lower than when gK splenocytes were stimulated with peptide #2 (Fig. 2). Similarly, peptide #2 stimulated lymphocytes from KOS-immunized mice, but no significant responses were detected when splenocytes from gD- or mock-immunized groups were stimulated with peptide # 2 (not shown). Since none of the other 32 peptides induced any CD4+IFN-γ+ or CD8+IFN-γ+ stimulation, only an example of two of the 32 peptides are shown in Fig. 2 (peptides #1 and #3). As expected, peptides #1 and #3 did not stimulate splenocytes from gK-immunized mice; this was similar to the response in the unstimulated group (Fig. 2). Finally, the level of CD8+IFN-γ+ cells produced following stimulation of gK splenocytes with ConA was similar to that of gK splenocytes stimulated with peptide #2, while the level of CD4+IFN-γ+ following stimulation with ConA was lower than when the gK splenocytes were stimulated with peptide #2 (Fig. 2). These results suggest that peptide #2 stimulated lymphocytes from gK-immunized mice and both CD4+ and CD8+ T cells are involved in production of IFN-γ, following stimulation with peptide #2.
Fig. 2. Generation of CD4+IFN-γ+ and CD8+IFN-γ+ T cells following stimulation of splenocytes from gK-immunized mice with peptide #2.
Mice were immunized with gK, three times as described in Materials and Methods. Three weeks after the third immunization, splenocytes form immunized mice were isolated, and 2 ×106 cells were stimulated with 10 μg of each peptide, 10 PFU/cell of UV-inactivated HSV-1 strain McKrae, 1 μg of ConA, or 10 μg of irrelevant peptide. Four hours before harvesting the cells for FACS analyses, BFA was added as described in Materials and Methods. CD4+ and CD8+ T cells expressing intracellular IFN-γ in response to stimulators were identified using appropriate Abs in a FACSCalibur. The frequency of CD4+ and CD8+ T cells expressing IFN-γ in one representative mouse from gK-immunized mice is shown. Experiments were repeated 2X.
IFN-γ production by T cells following stimulation with peptide #2 is CD8-dependent
The data presented in Fig. 2 and discussed above suggest that both CD4+ and CD8+ T cells produce IFN-γ. Thus, IFN-γ production by CD4+ and CD8+ T cells can be independently regulated by each T cell subtype or could be CD4- or CD8-dependent. To determine the contribution of CD4+ and CD8+ T cells to IFN-γ production following stimulation of each T cell subpopulation with peptide #2, splenocytes from gK-immunized or KOS-immunized mice were fractionated into CD4+-or CD8+-only populations as described in Materials and Methods. Fractionated CD4+ or CD8+ T cell subtypes were stimulated as above with 10 μg of peptide #2 (or peptide #1 as a negative control) for 72 hr. Subsequently, the level of IFN-γ secreted into the media was analyzed by ELISA (as described in Materials and Methods). As shown in Fig. 3, CD8+ T cells following stimulation with peptide #2, but not peptide #1, secreted high levels of IFN-γ (Fig. 3; p <0.0001). In contrast, CD4+ T cells did not secret any significant amount of IFN-γ following in vitro stimulation with peptide #2 or #1. These differences for IFN-γ production between CD4+ versus CD8+ T cells following stimulation with peptide #2 were highly significant (Fig. 3; p<0.0001). These results suggest that IFN-γ production by CD4+ T cells is CD8+ T cell-dependent.
Fig. 3. IFN-γ production by splenocytes of gK-immunized mice is CD8-dependent.
Splenocytes from gK-immunized or KOS-immunized mice were separated into CD8+ or CD4+ T cells, and 2 × 106 cells were stimulated with 10 μg of peptide #2 or control peptide #1 in vitro for 72 hr as described in Materials and Methods. The concentrations of IFN-γ in the supernatants were measured by ELISA. Each point represents mean ± SEM for three replicates from three mice.
Induction of CTL activity in vivo by peptide #2
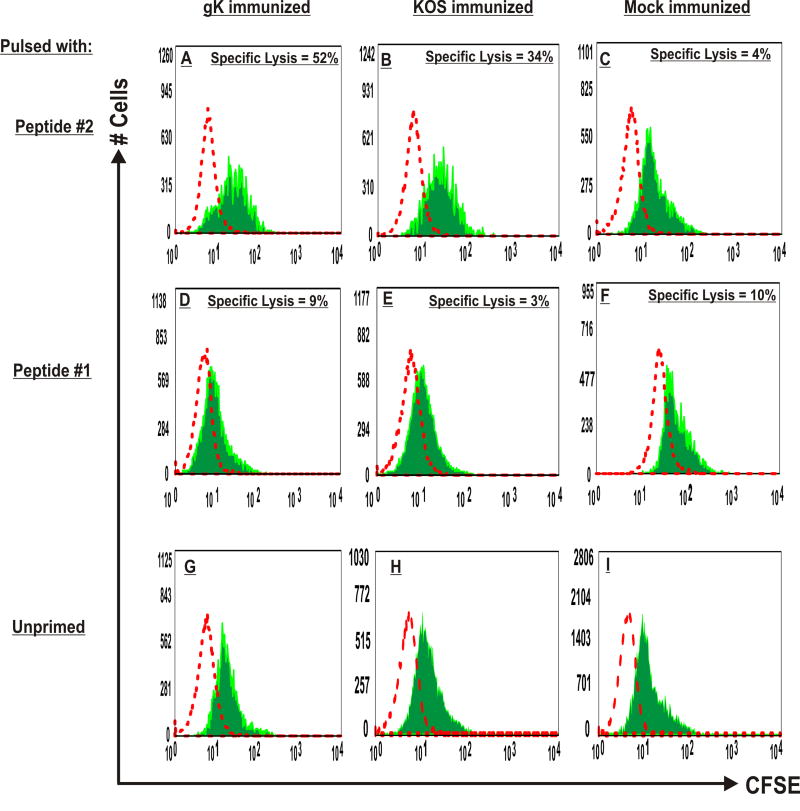
The cytolytic function of peptide #2-specific CD8+ T cells within the spleen of gK mice was determined using an in vivo cytotoxicity assay to measure the disappearance of CFSE-labeled, gK peptide #2-pulsed spleen cells as targets. Control mice were immunized with HSV-1 strain KOS or mock-immunized. T cells from gK-immunized mice that were pulsed with peptide #2 showed an approximately 52% lysis of target cells (Fig. 4A). Also, when KOS-immunized mice received T cells that were pulsed with peptide #2, they showed 34% cell lysis (Fig. 4B), but this was lower than the cell lysis detected in gK-immunized mice (Fig. 4A). The cell lysis in mock-immunized mice with lymphocytes pulsed with peptide #2 was 4% (Fig. 4C). In contrast, target cell lysis in gK-, KOS-, or mock-immunized mice with splenocytes stimulated with peptide #1 (Fig. 4; panels D, E, and F) or irrelevant VSV peptide (not shown) was <10%. Finally, when recipient gK- or KOS-immunized mice received lymphocytes that were pulsed with Group 1, Group 2, Group 3, or Group 4 peptides, they all induced cell lysis similarly to what was obtained with peptide #1 (not shown). This result suggests that peptide #2 has CTL activity in vivo.
Fig. 4. Induction of CTL activity in vivo by peptide #2.
To prepare target cells for detection of in vivo cytolytic activity, splenocytes from naive BALB/c mice were prepared as described in Materials and Methods. The cells were split into two populations. One population was pulsed with 10 μg of peptide #2, incubated at 37°C for 18 hr, and labeled with a high concentration of CFSE (2.5 μM) (CFSEhigh cells). The second control target population was pulsed with a nonspecific control VSV peptide and labeled with a low concentration of CFSE (0.25 μM) (CFSElow cells). For IV injection, an equal number of cells from each population were mixed together such that each mouse received one mouse’s worth of splenocytes in 100 μl of PBS. The target cells were then injected into mice that had previously been immunized with gK or KOS, or mock-immunized 3X as described in Materials and Methods. Four hours after IV injection of target cells, mice were sacrificed and their spleens were isolated. Cell suspensions were analyzed by FACS. Each population was detected by their differential CFSE fluorescence intensities. Representative histograms from various groups are shown. The specific lysis for each group was calculated as described in Materials and Methods using the intensity of low and high CFSE staining in relation to the unprimed control (panels G, H, and I). Panels: A) gK-immunized mice received lymphocytes primed with peptide #2; B) KOS-immunized mice received lymphocytes primed with peptide #2; C) Mock-immunized mice received lymphocytes primed with peptide #2; D) gK-immunized mice received lymphocytes primed with peptide #1; E) KOS-immunized mice received lymphocytes primed with peptide #1; F) Mock-immunized mice received lymphocytes primed with peptide #1; G) gK-immunized mice received unprimed lymphocytes; H) KOS-immunized mice received unprimed lymphocytes; and I) Mock-immunized mice received unprimed lymphocytes. Data are representative of FACS analyses from three mice per group.
Discussion
We demonstrated previously that immunization of mice with gK, but not with any of the other ten known HSV-1 glycoproteins, resulted in exacerbation of CS and herpetic dermatitis following ocular HSV-1 infection (Ghiasi et al., 1995; Ghiasi et al., 1994a). We also found that the exacerbation of CS in gK-immunized mice was due to the presence of CD8+ T cells in the cornea of ocularly infected mice (Osorio et al., 2004). Depletion of CD8+ T cells, rather than CD4+ T cells or macrophages, reduced the severity of the eye disease to the level of mock-immunized and ocularly infected mice. Using different protein antigens in conjunction with Freund’s Adjuvant, gK is the only antigen that induces CD8+ T cell response in the cornea of ocularly infected mice. However, CD8+ T cells in the cornea of gK-immunized mice are CD25+ (Ghiasi et al. unpublished).
The main goal of this study was to delineate the epitope(s) of gK that is (are) involved in T cell activities in vitro and in vivo. Thus, by using 33 overlapping peptides and four different expressed fragments of gK, we mapped a single region of gK that is involved in CTL activities in vivo, as well as T cell stimulation in vitro. This peptide had a five-amino-acid overlap with the two adjacent 15-mer peptides, neither of which showed any T cell activity in vitro or in vivo. Similarly, using 20 non-overlapping peptides, each 15 amino acids long, we mapped the gK peptide involved in CTL activity to the same region as we showed here for the 33 overlapping peptides (not shown). We also have shown that CD4+ and CD8+ T cells produced IFN-γ; however IFN-γ expression by CD4+ T cells was CD8+ T cell-dependent. These results are in line with our previous studies showing that exacerbation of CS and herpetic dermatitis in gK-immunized mice following ocular infection was due to the presence of CD8+ T cells in the cornea of ocularly infected mice (Osorio et al., 2004). In this regard, it has been shown that 4-1BB, which has preferential effects on the survival or expansion of CD8+ T cells, is required for the development of herpes stromal keratitis (HSK) (Koelle et al., 1994; Seo et al., 2003; Verjans et al., 1998). It also has been shown that corneal bottoms isolated from HSK patients had higher numbers of HSV-reactive CD8+ T cells than HSV-reactive CD4+ T cells (Maertzdorf et al., 2003).
We used in silico analysis of the 338 amino acids sequence of gK to identify putative T cell epitopes based on prediction of MHC-binding determinants. We identified four clusters of polypeptide regions that contain several overlapping binding frames. These clusters represent the polypeptide segments A3-R42, M101-C144, I214-L264, and C300-F307. However, except for the region corresponding to the A3-R42 segment, the other three regions did not have any functional activities in vitro or in vivo. Thus, based on our mapping studies, we focused on the amino-terminal polypeptide cluster A3-R42 that is located in a predicted long α-helix region (McGuffin et al., 2000). High scores for a number of human and mouse MHC class I molecules were obtained for the peptide STVVLITAYGLVLVW (S11-W25). With regards to interaction with human class II alleles, this peptide is predicted to bind 84% of the alleles. With regards to human class I analysis, this peptide is predicted to bind HLA-A*0201, A*03, and A*26. Furthermore, this peptide is also predicted to bind H-2Db, H-2Kd and H-2Ld. This is in line with our previous studies that gK immunization exacerbated eye diseases in both BALB/c (H-2d) and C57BL/6 mice (H-2b) (Ghiasi et al., 1997). The results confirm our in vivo and in vitro studies that both CD4+ and CD8+ T cells can broadly recognize this peptide. These data suggest that the sequence is located in a functionally relevant domain of the protein and therefore preserved among HSV species. In fact, the sequence ITAYGLVL (I16-L23) is identical to the gK HSV-2 (I17-L23) (Dolan et al., 1998; McGeoch et al., 1991; McGeoch et al., 1988).
Our previous studies suggested that gK immunization induces CD8+ T cells in ocularly infected mice (Osorio et al., 2004), while the present study suggests that the CD8+ CTL activity in vivo is associated with peptide #2. Epitopes presented by class I MHC molecules are generated in the multi-step ubiquitin/proteosome/TAP pathway (York and Rock, 1996). In this process, ubiquitin binds to cytoplasmic or nuclear proteins, proteosomes degrade this complex into peptides, and TAP transports the degraded peptides into the ER. Studies using insertion/deletion mutants of gK have shown that gK is a transmembrane protein (Foster and Kousoulas, 1999; Foster et al., 2004). Since gK is a cell surface protein and may not normally be present in the cytoplasm, the binding of the peptide could occur in the ER. However, the mapped gK peptide is located within the possible signal peptide of gK (Mo and Holland, 1997; Ramaswamy and Holland, 1992). While there is no direct evidence about the cleavage of gK signal sequence during infection, topology studies of infected cells revealed that gK signal sequence is cleaved during the course of virus infection (Foster et al., 2004).” The gK signal peptide is 30 amino acids long, and there is the possibility that after cleavage, it directly binds to MHC molecules. Our two-hybrid results suggest that gK binds to signal peptidase, which is also known as the minor histocompatibility complex (manuscript in preparation). In line with this observation, it has been shown that signal peptidase or some proteases can generate peptides that bind directly to class I MHC molecules in the ER, thus bypassing the cytoplasmic ubiquitin/proteosome/TAP pathway (Hammond et al., 1995; Henderson et al., 1993; Lee et al., 1996; Wei and Cresswell, 1992). Thus, presentation of gK to MHC molecules may be associated with the ubiquitin/proteosome/TAP pathway, signal peptidase, or both.
The identification of a CD4+ and CD8+ T cells epitope within the signal sequence of gK posses an interesting mechanistic problem in terms of how this epitope could be loaded onto MHC I and MHC II molecules for presentation to T cells. Normally, the cleaved signal peptides are the substrates for the signal peptidase complex in inner leaflet of the ER membrane, where the signal sequences undergo site-specific cleavage during the translocation of the pre-proteins (Flu HA, HSV gD, HIV-1 gp160) across the membrane channel (Nayak and Jabbar, 1989; Salzwedel et al., 1993; Salzwedel et al., 1998; Spear et al., 1978). It has been presumed that such cleavage step on the membrane would release the signal peptide from the mature proteins in the ER lumen. In recent years, there is evidence to suggest that retro-translocation of the proteins could occur by specific mechanisms delivering peptides or proteins to the cytosol from the ER membrane or lumen (Ackerman et al., 2005; McCracken and Brodsky, 2003; Tsai et al., 2002). Such a retro-translocation process is intrinsically capable of loading MHC molecules with signal peptides in the lumen of the ER (Cresswell et al., 2005). There are a few instances where this could be operational in the presentation of the signal peptides of HIV-1 gp160 and HCV to T cells (Borrow et al., 1997; Kowalski et al., 1996). The fact that the epitope within the gK signal peptide is also recognized by both T cell (CD4+ and CD8+) subsets suggests an intriguing possibility that a similar retro-translocation process might be involved in the generation of gK-specific T cell responses in vivo.
In summary, we have mapped a unique gK CTL epitope to a region of 15 amino acids located within the proposed signal sequence of gK. This gK epitope is involved in CTL activity in vivo and IFN-γ expression in vitro. However, in vitro expression of IFN-γ by CD4+ T cells is CD8+ T cell-dependent. Additional studies are required to ascertain the specific role of this epitope in exacerbation of ocular disease following HSV-1 infections.
Acknowledgments
This work was supported by Public Health Service grant EY13615 from the National Eye Institute and the Skirball Program in Molecular Ophthalmology to HG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackerman AL, Kyritsis C, Tampe R, Cresswell P. Access of soluble antigens to the endoplasmic reticulum can explain cross-presentation by dendritic cells. Nature Immunol. 2005;6(1):107–13. doi: 10.1038/ni1147. [DOI] [PubMed] [Google Scholar]
- Bond VC, Person S. Fine structure physical map locations of alterations that affect cell fusion in herpes simplex virus type 1. Virology. 1984;132(2):368–376. doi: 10.1016/0042-6822(84)90042-4. [DOI] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, Nelson JA, Gairin JE, Hahn BH, Oldstone MB, Shaw GM. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nature Med. 1997;3(2):205–11. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- Cresswell P, Ackerman AL, Giodini A, Peaper DR, Wearsch PA. Mechanisms of MHC class I-restricted antigen processing and cross-presentation. Immunologi Rev. 2005;207:145–57. doi: 10.1111/j.0105-2896.2005.00316.x. [DOI] [PubMed] [Google Scholar]
- Debroy C, Pederson N, Person S. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology. 1985;145(1):36–48. doi: 10.1016/0042-6822(85)90199-0. [DOI] [PubMed] [Google Scholar]
- Dietz P, Klupp BG, Fuchs W, Kollner B, Weiland E, Mettenleiter TC. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J Virol. 2000;74(11):5083–90. doi: 10.1128/jvi.74.11.5083-5090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan A, Jamieson FE, Cunningham C, Barnett BC, McGeoch DJ. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72(3):2010–21. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TP, Alvarez X, Kousoulas KG. Plasma membrane topology of syncytial domains of herpes simplex virus type 1 glycoprotein K (gK): the UL20 protein enables cell surface localization of gK but not gK-mediated cell-to-cell fusion. J Virol. 2003;77(1):499–510. doi: 10.1128/JVI.77.1.499-510.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TP, Kousoulas KG. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J Virol. 1999;73(10):8457–68. doi: 10.1128/jvi.73.10.8457-8468.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TP, Melancon JM, Baines JD, Kousoulas KG. The herpes simplex virus type 1 UL20 protein modulates membrane fusion events during cytoplasmic virion morphogenesis and virus-induced cell fusion. J Virol. 2004;78(10):5347–57. doi: 10.1128/JVI.78.10.5347-5357.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiasi H, Bahri S, Nesburn AB, Wechsler SL. Protection against herpes simplex virus-induced eye disease after vaccination with seven individually expressed herpes simplex virus 1 glycoproteins. Invest Ophthalmol Vis Sci. 1995;36(7):1352–1360. [PubMed] [Google Scholar]
- Ghiasi H, Cai S, Slanina S, Nesburn AB, Wechsler SL. Nonneutralizing antibody against the glycoprotein K of herpes simplex virus type-1 exacerbates herpes simplex virus type-1-induced corneal scarring in various virus-mouse strain combinations. Invest Ophthalmol Vis Sci. 1997;38(6):1213–21. [PubMed] [Google Scholar]
- Ghiasi H, Kaiwar R, Nesburn AB, Slanina S, Wechsler SL. Expression of seven herpes simplex virus type 1 glycoproteins (gB, gC, gD, gE, gG, gH, and gI): comparative protection against lethal challenge in mice. J Virol. 1994a;68(4):2118–2126. doi: 10.1128/jvi.68.4.2118-2126.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiasi H, Slanina S, Nesburn AB, Wechsler SL. Characterization of baculovirus-expressed herpes simplex virus type 1 glycoprotein K. J Virol. 1994b;68(4):2347–2354. doi: 10.1128/jvi.68.4.2347-2354.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P, Doytchinova IA, Zygouri C, Flower DR. MHCPred: bringing a quantitative dimension to the online prediction of MHC binding. Applied Bioinformatics. 2003;2(1):63–6. [PubMed] [Google Scholar]
- Hammond SA, Johnson RP, Kalams SA, Walker BD, Takiguchi M, Safrit JT, Koup RA, Siliciano RF. An epitope-selective, transporter associated with antigen presentation (TAP)-1/2-independent pathway and a more general TAP-1/2-dependent antigen-processing pathway allow recognition of the HIV-1 envelope glycoprotein by CD8+ CTL. J Immunol. 1995;154(11):6140–56. [PubMed] [Google Scholar]
- Henderson RA, Cox AL, Sakaguchi K, Appella E, Shabanowitz J, Hunt DF, Engelhard VH. Direct identification of an endogenous peptide recognized by multiple HLA-A2.1-specific cytotoxic T cells. Proc Nat Acad of Sci (USA) 1993;90(21):10275–9. doi: 10.1073/pnas.90.21.10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson L, Goldsmith K, Snoddy D, Ghosh H, Graham FL, Johnson DC. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J Virol. 1992;66(9):5603–5609. doi: 10.1128/jvi.66.9.5603-5609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson L, Johnson DC. Herpes simplex virus glycoprotein K promotes egress of virus particles. J Virol. 1995;69(9):5401–5413. doi: 10.1128/jvi.69.9.5401-5413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson L, Roop-Beauchamp C, Johnson DC. Herpes simplex virus glycoprotein K is known to influence fusion of infected cells, yet is not on the cell surface. J Virol. 1995;69(7):4556–4563. doi: 10.1128/jvi.69.7.4556-4563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassim SH, Rajasagi NK, Zhao X, Chervenak R, Jennings SR. In vivo ablation of CD11c-positive dendritic cells increases susceptibility to herpes simplex virus type 1 infection and diminishes NK and T-cell responses. J Virol. 2006;80(8):3985–93. doi: 10.1128/JVI.80.8.3985-3993.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klupp BG, Baumeister J, Dietz P, Granzow H, Mettenleiter TC. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J Virol. 1998;72(3):1949–58. doi: 10.1128/jvi.72.3.1949-1958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle DM, Abbo H, Peck A, Ziegweid K, Corey L. Direct recovery of herpes simplex virus (HSV)-specific T lymphocyte clones from recurrent genital HSV-2 lesions. J Infect Dis. 1994;169(5):956–961. doi: 10.1093/infdis/169.5.956. [DOI] [PubMed] [Google Scholar]
- Kowalski H, Erickson AL, Cooper S, Domena JD, Parham P, Walker CM. Patr-A and B, the orthologues of HLA-A and B, present hepatitis C virus epitopes to CD8+ cytotoxic T cells from two chronically infected chimpanzees. J Exp Med. 1996;183(4):1761–75. doi: 10.1084/jem.183.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SP, Thomas WA, Blake NW, Rickinson AB. Transporter (TAP)-independent processing of a multiple membrane-spanning protein, the Epstein-Barr virus latent membrane protein 2. Eur J Immunol. 1996;26(8):1875–83. doi: 10.1002/eji.1830260831. [DOI] [PubMed] [Google Scholar]
- Little SP, Schaffer PA. Expression of the syncytial (syn) phenotype in HSV-1, strain KOS: genetic and phenotypic studies of mutants in two syn loci. Virology. 1981;112(2):686–702. doi: 10.1016/0042-6822(81)90314-7. [DOI] [PubMed] [Google Scholar]
- Maertzdorf J, Verjans GM, Remeijer L, van der Kooi A, Osterhaus AD. Restricted T cell receptor beta-chain variable region protein use by cornea-derived CD4+ and CD8+ herpes simplex virus-specific T cells in patients with herpetic stromal keratitis. J Infect Dis. 2003;187(4):550–8. doi: 10.1086/367991. [DOI] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL. Evolving questions and paradigm shifts in endoplasmic-reticulum-associated degradation (ERAD) Bioessays. 2003;25(9):868–77. doi: 10.1002/bies.10320. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Cunningham C, McIntyre G, Dolan A. Comparative sequence analysis of the long repeat regions and adjoining parts of the long unique regions in the genomes of herpes simplex viruses types 1 and 2. J Gen Virol. 1991;72(Pt 12):3057–3075. doi: 10.1099/0022-1317-72-12-3057. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Dalrymple MA, Davison AJ, Dolan A, Frame MC, McNab D, Perry LJ, Scott JE, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69(Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16(4):404–5. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Mo C, Holland TC. Determination of the transmembrane topology of herpes simplex virus type 1 glycoprotein K. J Biol Chem. 1997;272(52):33305–11. doi: 10.1074/jbc.272.52.33305. [DOI] [PubMed] [Google Scholar]
- Mo C, Suen J, Sommer M, Arvin A. Characterization of Varicella-Zoster virus glycoprotein K (open reading frame 5) and its role in virus growth. J Virol. 1999;73(5):4197–207. doi: 10.1128/jvi.73.5.4197-4207.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa AS, Shaban FA. ProPred analysis and experimental evaluation of promiscuous T-cell epitopes of three major secreted antigens of Mycobacterium tuberculosis. Tuberculosis. 2006;86(2):115–24. doi: 10.1016/j.tube.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Nayak DP, Jabbar MA. Structural domains and organizational conformation involved in the sorting and transport of influenza virus transmembrane proteins. Ann Rev Microbiol. 1989;43:465–501. doi: 10.1146/annurev.mi.43.100189.002341. [DOI] [PubMed] [Google Scholar]
- Osorio Y, Cai S, Hofman FM, Brown DJ, Ghiasi H. Involvement of CD8+ T cells in exacerbation of corneal scarring in mice. Curr Eye Res. 2004;29:145–151. doi: 10.1080/02713680490504632. [DOI] [PubMed] [Google Scholar]
- Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152(1):163–75. [PubMed] [Google Scholar]
- Pogue-Geile KL, Spear PG. The single base pair substitution responsible for the Syn phenotype of herpes simplex virus type 1, strain MP. Virology. 1987;157(1):67–74. doi: 10.1016/0042-6822(87)90314-x. [DOI] [PubMed] [Google Scholar]
- Ramaswamy R, Holland TC. In vitro characterization of the HSV-1 UL53 gene product. Virology. 1992;186(2):579–587. doi: 10.1016/0042-6822(92)90024-j. [DOI] [PubMed] [Google Scholar]
- Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(34):213–9. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- Reche PA, Glutting JP, Zhang H, Reinherz EL. Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics. 2004;56(6):405–19. doi: 10.1007/s00251-004-0709-7. [DOI] [PubMed] [Google Scholar]
- Salzwedel K, Johnston PB, Roberts SJ, Dubay JW, Hunter E. Expression and characterization of glycophospholipid-anchored human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1993;67(9):5279–88. doi: 10.1128/jvi.67.9.5279-5288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzwedel K, West JT, Jr, Mulligan MJ, Hunter E. Retention of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum does not redirect virus assembly from the plasma membrane. J Virol. 1998;72(9):7523–31. doi: 10.1128/jvi.72.9.7523-7531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo SK, Park HY, Choi JH, Kim WY, Kim YH, Jung HW, Kwon B, Lee HW, Kwon BS. Blocking 4-1BB/4-1BB ligand interactions prevents herpetic stromal keratitis. J Immunol. 2003;171(2):576–83. doi: 10.4049/jimmunol.171.2.576. [DOI] [PubMed] [Google Scholar]
- Singh H, Raghava GP. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17(12):1236–7. doi: 10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- Spear PG, Sarmiento M, Manservigi R. The structural proteins and glycoproteins of herpesviruses: a review. IARC Sci Publ. 1978;24:157–167. [PubMed] [Google Scholar]
- Tsai B, Ye Y, Rapoport TA. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nature Rev Mol Cell Biol. 2002;3(4):246–55. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- Verjans GM, Feron EJ, Dings ME, Cornelissen JG, Van der Lelij A, Baarsma GS, Osterhaus AD. T cells specific for the triggering virus infiltrate the eye in patients with herpes simplex virus-mediated acute retinal necrosis. J Infect Dis. 1998;178(1):27–34. doi: 10.1086/515586. [DOI] [PubMed] [Google Scholar]
- Wei ML, Cresswell P. HLA-A2 molecules in an antigen-processing mutant cell contain signal sequence-derived peptides. Nature. 1992;356(6368):443–6. doi: 10.1038/356443a0. [DOI] [PubMed] [Google Scholar]
- York IA, Rock KL. Antigen processing and presentation by the class I major histocompatibility complex. Ann Rev Immunol. 1996;14:369–96. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]