Abstract
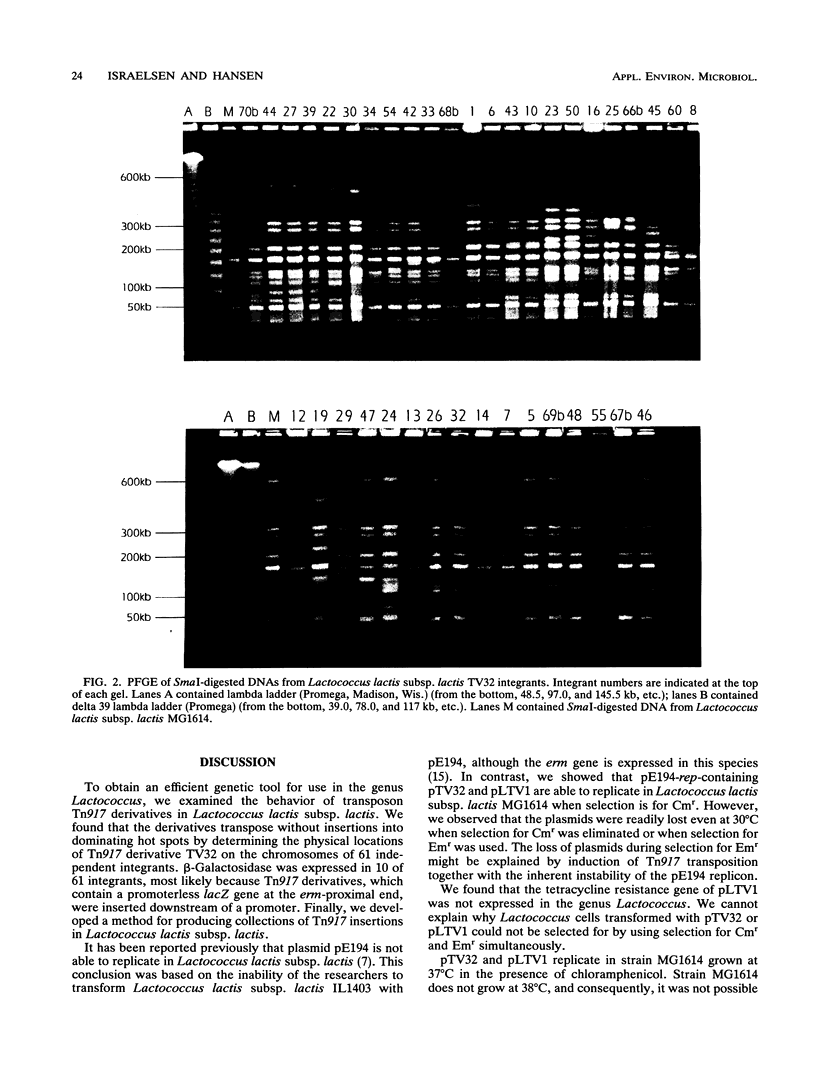
Two transposition vectors, pTV32 and pLTV1, containing transposon Tn917 derivatives TV32 and LTV1, respectively, were introduced into Lactococcus lactis subsp. lactis MG1614. It was found that pTV32 and pLTV1 replicate and that TV32 and LTV1 transpose in this strain. A protocol for production of a collection of Tn917 insertions in L. lactis subsp. lactis was developed. The physical locations of TV32 on the chromosomal SmaI fragments of 62 independent transpositions were established by pulsed-field gel electrophoresis. These transpositions could be divided into at least 38 different groups that exhibited no Tn917-dominating hot spots on the L. lactis subsp. lactis chromosome. A total of 10 of the 62 transpositions resulted in strains that express β-galactosidase. This indicates that there was fusion of the promoterless lacZ of the Tn917 derivatives to a chromosomal promoter. Thus, the Tn917-derived transposons should be powerful genetic tools for studying L. lactis subsp. lactis.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boe L., Nielsen T. T., Madsen S. M., Andrup L., Bolander G. Cloning and characterization of two plasmids from Bacillus thuringiensis in Bacillus subtilis. Plasmid. 1991 May;25(3):190–197. doi: 10.1016/0147-619x(91)90012-l. [DOI] [PubMed] [Google Scholar]
- Bohall N. A., Jr, Vary P. S. Transposition of Tn917 in Bacillus megaterium. J Bacteriol. 1986 Aug;167(2):716–718. doi: 10.1128/jb.167.2.716-718.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli A., Portnoy A., Youngman P. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J Bacteriol. 1990 Jul;172(7):3738–3744. doi: 10.1128/jb.172.7.3738-3744.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin M. C., Chopin A., Rouault A., Galleron N. Insertion and amplification of foreign genes in the Lactococcus lactis subsp. lactis chromosome. Appl Environ Microbiol. 1989 Jul;55(7):1769–1774. doi: 10.1128/aem.55.7.1769-1774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos W. M., Simons G. Molecular cloning of lactose genes in dairy lactic streptococci: the phospho-beta-galactosidase and beta-galactosidase genes and their expression products. Biochimie. 1988 Apr;70(4):461–473. doi: 10.1016/0300-9084(88)90083-1. [DOI] [PubMed] [Google Scholar]
- Froseth B. R., McKay L. L. Molecular characterization of the nisin resistance region of Lactococcus lactis subsp. lactis biovar diacetylactis DRC3. Appl Environ Microbiol. 1991 Mar;57(3):804–811. doi: 10.1128/aem.57.3.804-811.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holo H., Nes I. F. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol. 1989 Dec;55(12):3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R. Bacteriocins of lactic acid bacteria. Biochimie. 1988 Mar;70(3):337–349. doi: 10.1016/0300-9084(88)90206-4. [DOI] [PubMed] [Google Scholar]
- Kok J. Genetics of the proteolytic system of lactic acid bacteria. FEMS Microbiol Rev. 1990 Sep;7(1-2):15–42. doi: 10.1111/j.1574-6968.1990.tb04877.x. [DOI] [PubMed] [Google Scholar]
- Kok J., van der Vossen J. M., Venema G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984 Oct;48(4):726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourgeois P., Mata M., Ritzenthaler P. Genome comparison of Lactococcus strains by pulsed-field gel electrophoresis. FEMS Microbiol Lett. 1989 May;50(1-2):65–69. doi: 10.1016/0378-1097(89)90460-6. [DOI] [PubMed] [Google Scholar]
- Mayo B., Kok J., Venema K., Bockelmann W., Teuber M., Reinke H., Venema G. Molecular cloning and sequence analysis of the X-prolyl dipeptidyl aminopeptidase gene from Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1991 Jan;57(1):38–44. doi: 10.1128/aem.57.1.38-44.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi M., Chopin M. C., Chopin A., Cals M. M., Gripon J. C. Cloning and DNA sequence analysis of an X-prolyl dipeptidyl aminopeptidase gene from Lactococcus lactis subsp. lactis NCDO 763. Appl Environ Microbiol. 1991 Jan;57(1):45–50. doi: 10.1128/aem.57.1.45-50.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins J. B., Youngman P. J. A physical and functional analysis of Tn917, a Streptococcus transposon in the Tn3 family that functions in Bacillus. Plasmid. 1984 Sep;12(2):119–138. doi: 10.1016/0147-619x(84)90058-1. [DOI] [PubMed] [Google Scholar]
- Sanders M. E., Nicholson M. A. A method for genetic transformation of nonprotoplasted Streptococcus lactis. Appl Environ Microbiol. 1987 Aug;53(8):1730–1736. doi: 10.1128/aem.53.8.1730-1736.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E. Phage resistance in lactic acid bacteria. Biochimie. 1988 Mar;70(3):411–422. doi: 10.1016/0300-9084(88)90215-5. [DOI] [PubMed] [Google Scholar]
- Shaw J. H., Clewell D. B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985 Nov;164(2):782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanskanen E. I., Tulloch D. L., Hillier A. J., Davidson B. E. Pulsed-Field Gel Electrophoresis of SmaI Digests of Lactococcal Genomic DNA, a Novel Method of Strain Identification. Appl Environ Microbiol. 1990 Oct;56(10):3105–3111. doi: 10.1128/aem.56.10.3105-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomich P. K., An F. Y., Clewell D. B. Properties of erythromycin-inducible transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1980 Mar;141(3):1366–1374. doi: 10.1128/jb.141.3.1366-1374.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeyar M. A., Zahler S. A. Chromosomal insertions of Tn917 in Bacillus subtilis. J Bacteriol. 1986 Aug;167(2):530–534. doi: 10.1128/jb.167.2.530-534.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P. J., Perkins J. B., Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P., Zuber P., Perkins J. B., Sandman K., Igo M., Losick R. New ways to study developmental genes in spore-forming bacteria. Science. 1985 Apr 19;228(4697):285–291. doi: 10.1126/science.228.4697.285. [DOI] [PubMed] [Google Scholar]
- van Belkum M. J., Hayema B. J., Jeeninga R. E., Kok J., Venema G. Organization and nucleotide sequences of two lactococcal bacteriocin operons. Appl Environ Microbiol. 1991 Feb;57(2):492–498. doi: 10.1128/aem.57.2.492-498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]