Abstract
Background
The evolutionary lineage leading to the teleost fish underwent a whole genome duplication termed FSGD or 3R in addition to two prior genome duplications that took place earlier during vertebrate evolution (termed 1R and 2R). Resulting from the FSGD, additional copies of genes are present in fish, compared to tetrapods whose lineage did not experience the 3R genome duplication. Interestingly, we find that ParaHox genes do not differ in number in extant teleost fishes despite their additional genome duplication from the genomic situation in mammals, but they are distributed over twice as many paralogous regions in fish genomes.
Results
We determined the DNA sequence of the entire ParaHox C1 paralogon in the East African cichlid fish Astatotilapia burtoni, and compared it to orthologous regions in other vertebrate genomes as well as to the paralogous vertebrate ParaHox D paralogons. Evolutionary relationships among genes from these four chromosomal regions were studied with several phylogenetic algorithms. We provide evidence that the genes of the ParaHox C paralogous cluster are duplicated in teleosts, just as it had been shown previously for the D paralogon genes. Overall, however, synteny and cluster integrity seems to be less conserved in ParaHox gene clusters than in Hox gene clusters. Comparative analyses of non-coding sequences uncovered conserved, possibly co-regulatory elements, which are likely to contain promoter motives of the genes belonging to the ParaHox paralogons.
Conclusion
There seems to be strong stabilizing selection for gene order as well as gene orientation in the ParaHox C paralogon, since with a few exceptions, only the lengths of the introns and intergenic regions differ between the distantly related species examined. The high degree of evolutionary conservation of this gene cluster's architecture in particular – but possibly clusters of genes more generally – might be linked to the presence of promoter, enhancer or inhibitor motifs that serve to regulate more than just one gene. Therefore, deletions, inversions or relocations of individual genes could destroy the regulation of the clustered genes in this region. The existence of such a regulation network might explain the evolutionary conservation of gene order and orientation over the course of hundreds of millions of years of vertebrate evolution. Another possible explanation for the highly conserved gene order might be the existence of a regulator not located immediately next to its corresponding gene but further away since a relocation or inversion would possibly interrupt this interaction. Different ParaHox clusters were found to have experienced differential gene loss in teleosts. Yet the complete set of these homeobox genes was maintained, albeit distributed over almost twice the number of chromosomes. Selection due to dosage effects and/or stoichiometric disturbance might act more strongly to maintain a modal number of homeobox genes (and possibly transcription factors more generally) per genome, yet permit the accumulation of other (non regulatory) genes associated with these homeobox gene clusters.
Background
The cichlid fish
Cichlids belong to the most diverse and species-rich families of fishes. With an estimated number of more than 3,000 species they alone represent more than ten percent of all fish species. The family Cichlidae belongs to the teleosts that, with more than 26,500 species, are the most diverse lineage of all vertebrates [1]. Cichlids have a Gondwanian distribution and are found in India, Madagascar, South and Central America and Africa and developed a stunning variety of coloration patterns, body shapes, behaviors and trophic as well as ecological specializations within a few millions of years see [2-8]. Their unparalleled diversity made the cichlid species flocks a textbook example for parallel adaptive radiations and explosive speciation [7].
The evolutionary success of the cichlids has been attributed to morphological and behavioral patterns, although the relative importance of different mechanisms – as there will be surely more than one – is still debated. One plausible factor that is at least partly responsible for the cichlids' unique diversity is the complexity of their breeding system and social behavior. Cichlids evolved a variety of brood care strategies and mating systems, and it is likely that female choice with respect to male coloration played an important role during cichlid evolution [2,5,9-11]. Another possible reason for their evolutionary success is the particular architecture of the cichlids' jaw apparatus. They possess two sets of jaws, one oral and one pharyngeal jaw derived from the fifth gill arch. These jaws evolved independently from each other and allow for an immense variety of possible feeding types leading to different diets. Therefore, many different niches could be colonized by cichlids [12]. There is a large amount of behavioral and morphological divergence between different cichlid species in the East African lakes. Yet, rather surprising parallelisms have evolved in species flocks of the different lakes [3,5,8] indicating that the genetic "predisposition" for the modification of these traits might have been already present in the genome of the common ancestor of all the East African cichlid species. We assume that a substantial part of the necessary modifications of the cichlids' genome takes place in the regulatory elements of only a few important genes. To test this hypothesis it would be important to identify those genes of relevance in speciation. As part of this overall research effort we focus here on the ParaHox genes, a sister-cluster of the Hox genes that are crucial in development [13,14]. Here, we report on an investigation of the genomics of the ParaHox C and D paralogons of the cichlid Astatotilapia burtoni and present the results of a comparison of some of its genomic features with those of other vertebrate ParaHox clusters.
Genome duplication, Hox- and ParaHox clusters in vertebrates
It has been suggested that gene- or genome duplications might be important evolutionary mechanisms resulting in new copies of genes, which are then free to accumulate mutations and to evolve new or additional functions [15]. Changes in regulatory elements of duplicated gene copies could, for example, cause neofunctionalization; the gain of a new function, or a subfunctionalization; i.e., subdividing the original functions of the duplicated gene between the daughter genes [16]. Genes under relaxed selection can arise after the duplication of single genes, large chromosomal fragments or even whole genomes [[17] and references therein]. For each of these three possibilities, different effects are characteristic: the preservation or disruption of regulatory control, the genomic context, the potential for dosage imbalance and, of course, the size of the duplicated fragment [18].
Duplications of genes as a consequence of the activity of transposable elements, unequal crossing-over and other mechanisms occur frequently in the course of vertebrate evolution [19]. The duplication of whole genomes, however is a rare event in animals, although there are quite a few polyploid species in some taxonomic groups such as frogs [20], salamanders [21] and several fish lineages such as salmonids [22], cyprinids and catfish [23]). In plants polyploidy is a rather common phenomenon [24-26].
Several studies have proposed the existence of two rounds of whole genome duplications during vertebrate evolution (2R hypothesis) [[14,27] and references therein]. More recent analyses revealed that in the lineage leading to the ray-finned fish, an additional genome duplication event, the fish-specific genome duplication (3R or FSGD), has occurred [28-33]. The 1R and 2R can be roughly dated 430 – 750 mya [27,34] in the lineage of the Gnathostomata. However, the phylogenetic relationships of the agnathan lineages to one other and to the vertebrates as well the timing of 1R and 2R is not fully resolved yet [35]. The FSGD [36,37] took place in the lineage of ray-finned fish, after the separation of gars but before the origin of the Osteoglossomorpha [30], around 320 mya [27,38].
Among the first to be discovered and still among the most prominent examples for duplicated genes through whole genome duplications are the Hox clusters [14]. The number of Hox gene clusters and their genomic architecture in vertebrate genomes are an excellent illustration for the vertebrate genomic history of two rounds of genome duplications (1R, 2R), as well as an additional fish specific genome duplication (3R/FSGD) [39]. One cluster is found in the genome of the Cephalochordate Branchiostoma and one cluster is assumed to be the ancestral state [40]. Two rounds of genome duplication led to four copies in sharks and tetrapods and another round of genome duplication along with reciprocal losses of genes lead to a total number seven Hox clusters in teleost fish [33,39]. Therefore it might be expected that the genes of the ParaHox clusters, just as those of the Hox clusters, should reflect the history of the last two genome duplications in fish as well [41].
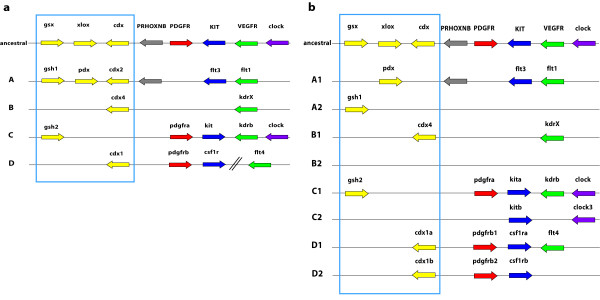
The ParaHox complex in mammals consists, just as the Hox complex, out of four clusters termed A to D. But only the A cluster still carries all three genes of the predicted ancestral ParaHox cluster. All other clusters contain only a single ParaHox gene at most. The ParaHox complex is even more fragmented in teleost fish, and Mulley et al. [42] argued that there are no teleost ParaHox clusters at all. However, if the adjoining genes of the ParaHox clusters are taken into account, the syntenic structure of the genomic region/paralogon that contains the ParaHox gene(s) becomes apparent [43-45] (see Figure 1) and the evolutionary history of the clusters can be reconstructed.
Figure 1.
Schematic of the genomic architecture of the vertebrate ParaHox loci. a: Structure of the presumed ancestral vertebrate condition and the mammalian ParaHox clusters and their 3' adjoining genes ("ParaHox paralogon"). b: structure of the derived (post FSGD) teleost fish ParaHox cluster genomic architecture and their 3' adjacent genes [43-45] The color code of the relevant genes is maintianed throughout this paper: yellow: ParaHox genes, red: pdgfr genes, blue: kit and csf1r genes, green: vegfr genes, purple: clock genes; the colors red, blue and green also stand for RTKs type III.
The present study regards a ParaHox paralogon as the ParaHox gene(s) from a cluster together with the respective 3' adjoining genes as a [46] (see Figure 1). Therefore, genes located 5'of the whole paralogon are referred to as 5'of a gene X and genes more towards the 3'end of a paralogon are referred to as 3'of a particular gene X, irrespective of the orientation of gene X.
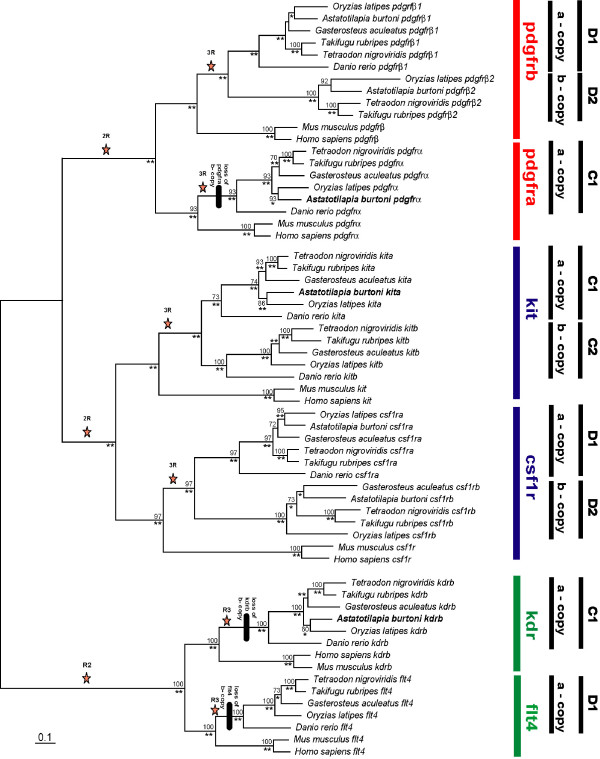
For this study we conducted an analysis of the genomic evolution of vertebrate ParaHox paralogons. Specifically, we were interested in the ParaHox clusters C and D and the adjoining type III receptor tyrosine kinase genes (pdgfrα/kit and pdgfrβ/csf1r respectively) that were shown to be involved in teleost coloration [46-49]. To this end, we determined the DNA sequence of the entire ParaHox C1 paralogon in the East African cichlid fish Astatotilapia burtoni, and compared it to orthologous regions in other teleosts' genomes (Danio rerio, Oryzias latipes, Tetraodon nigroviridis, Takifugu rubripes, Gasterosteus aculeatus) and mammals (Mus musculus, Homo sapiens) as well as to the Astatotilapia burtoni ParaHox D paralogons [46]. This was done in order to investigate the genomic consequences of several rounds of genome duplication in the vertebrate lineage (Figure 2).
Figure 2.
Phylogenetic analysis of the RTKs of the C and D ParaHox paralogon. Bayesian analysis, **: 1.00; *: 0.95 – 0.99 support, nucleotide data, stars mark the postulated phylogenetic timing of whole genome duplications. Tree are derived from a PHYML analysis.
Results and discussion
To investigate the evolution of the vertebrate ParaHox paralogons C and D we shotgun sequenced a BAC clone of a BAC library of the East African cichlid fish Astatotilapia burtoni [50] that contained the C1 ParaHox paralogon, i.e., the ParaHox gene gsh2 and its 3'adjoining genes. The obtained BAC contig (GenBank accession EF526075 GenBank accession number: sequence will be submitted upon acceptance of the paper) was then further analyzed and compared to the sequences of two other BAC clones of the African cichlid Astatotilapia burtoni, 20D21 (DQ386647) and 26M7 (DQ386648) containing the D1 and D2 paralogons [46].
Sequence assembly and analysis
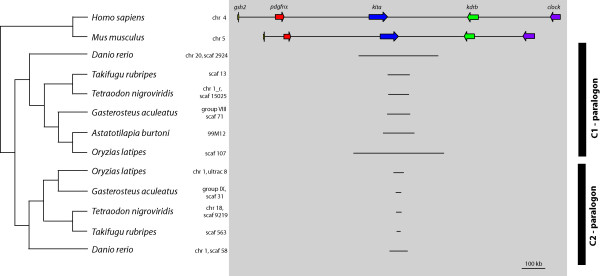
This analysis showed that the C(1) ParaHox gene locus and its 3' adjoining genes of Danio rerio is located on chromosome 20, of Takifugu rubripes on scaffold 13, of Tetraodon nigroviridis on 'chromosome1_random', of Gasterosteus aculeatus on group VIII, of Homo sapiens on chromosome 4, of Mus musculus of chromosome 5 and of Oryzias latipes on the scaffolds 1,264 (gsh2, pdgfrα), 578 (kita, kdrb) and 2,436 (clock) (see Figure 3 and see Additional File 1 [Table S1] for details).
Figure 3.
C1 and C2 ParaHox paralogon – overview. The C ParaHox paralogon of Homo sapiens, Tetraodon nigroviridis, Takifugu rubripes, Oryzias latipes, Danio rerio, Gasterosteus aculeatus and Astatotilapia burtoni. All teleost fishes except Astatotilapia are represented with their a- and b-copy. Of Astatotilapia only the a-copy is available at present. The orientation of the five genes and their respective paralogs are printed above the genes. The locations of the genes in their respective genome annotation are written on the right side of the name of the species.
Sequences for the C2 and the D ParaHox gene loci and their 3' adjoining genes were also retrieved from the aforementioned databases and aligned by hand. The locations of the different genes in the respective genome assemblies are summarized in Additional File 1.
Identification and characterization of Astatotilapia burtoni ParaHox paralogon containing BAC clones
The BAC library was screened for the C1 ParaHox paralogon gene kita as described previously [50]. A PCR screen for the presence of the ParaHox gene gsh2 was subsequently performed to identify BAC clones covering the entire C1 ParaHox paralogon. The kita and gsh2 positive clone 99M12, which was determined to have an insert length of 154 kb, was chosen for further investigation. The BAC clone was shotgun sequenced and BAC contigs were assembled into a scaffold and a complete sequence as described earlier [50].
BLAST searches [51] of the assembled contigs against GENBANK [52] showed that five genes were at least partially present in the BAC clone 99M12 (Figures 1 and 4): genomic screened homeo box 2 (gsh2), the platelet-derived growth factor receptor α (pdgfrα), the a – copy of the v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (kita), the b – copy of the kinase insert domain receptor (kdrb) and the circadian locomoter output cycles kaput (clock) (Figures 1 and 4). The C1 ParaHox paralogon was found to be the paralogon with the highest number of 3' adjoining genes in teleosts.
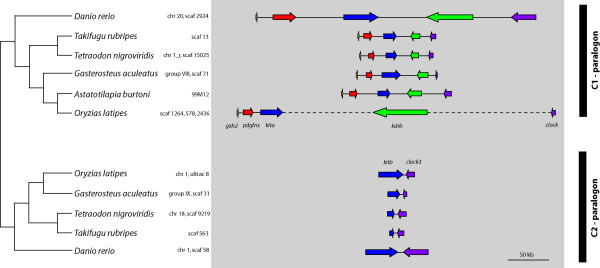
Figure 4.
C1 and C2 ParaHox paralogon. Detailed depiction of the teleost fish C1 and C2 ParaHox paralogons of Figure 1b. Homo sapiens and Mus musculus were left out of the figure to ensure a clear image of the genes. The orientation of the genes is indicated by the arrows.
Using cDNAs, annotated and predicted genes of Homo sapiens, Mus musculus and Danio rerio available on NCBI [52], we deduced the coding sequences of Takifugu rubripes, Tetraodon nigroviridis, Oryzias latipes, Gasterosteus aculeatus and Astatotilapia burtoni. We were able to assemble the complete coding sequences of four of the five genes located on the BAC clone 99M12 of A. burtoni. The only incompletely assembled gene is kdrb where approximately 200 bp of the coding sequence are missing.
From the beginning of the gene gsh2 to the end of clock this sequence of the clone 99M12 spans 133.56 kb. This length was used for comparisons of the lengths of the C1 ParaHox paralogons of the different organisms used in this study (Homo sapiens, Danio rerio, Takifugu rubripes, Tetraodon nigroviridis, Oryzias latipes, Gasterosteus aculeatus and Astatotilapia burtoni) because the real length of the inserted gaps is unknown as of present.
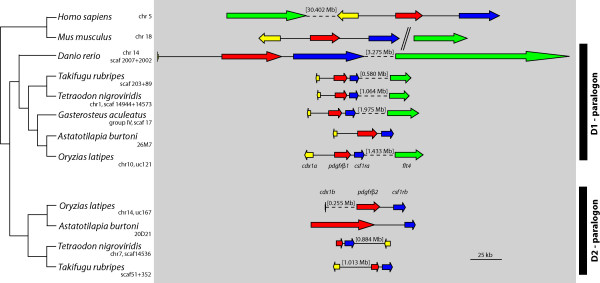
Another BAC clone (26M7) of the Astatotilapia burtoni BAC library contains the genes caudal type homeo box transcription factor 1 a (cdx1a), the platelet-derived growth factor receptor β1 (pdgfrβ1) and the colony-stimulating factor 1 receptor a (csf1ra) [46]. These three genes plus the fms-related tyrosine kinase 4 (flt4) that is not present on the clone belong to the D1 ParaHox paralogon, whereas the clone 20D21 was found to contain the genes platelet-derived growth factor receptor β2 (pdgfrβ2) and the colony-stimulating factor 1 receptor b (csf1rb) [46]. These two genes plus the caudal type homeobox transcription factor 1b (cdx1b) (not contained on this BAC clone) form the D2 ParaHox paralogon. The genes kitb and clock3, belong to the C2 ParaHox paralogon. Figure 5 shows the D1 and D2 ParaHox paralogons as defined above.
Figure 5.
D1 and D2 ParaHox paralogon. The D ParaHox paralogon of Homo sapiens, Mus musculus, Tetraodon nigroviridis, Takifugu rubripes, Oryzias latipes, Gasterosteus aculeatus and Astatotilapia burtoni. The orientations of the genes are indicated by the arrows. The chromosomal locations of these paralogons in their respective genomes are indicated next to the species name.
The ancestral ParaHox complex fish is fragmented in teleosts fish [42]. Therefore, we use the expression ParaHox "paralogon" instead of "cluster" since, especially for the case of the C2 ParaHox paralogon, not a single ParaHox gene is still present and in the case of the D2 ParaHox paralogon, the data we investigated did not include the ParaHox cdx1b gene. Possibly the most interesting finding is that the ParaHox complex of teleost fish, even after another round of whole genome duplication (WGD), the FSGD, and subsequent deletion of genes, contains exactly the same number of genes and orthologous set of ParaHox genes as the mammalian four ParaHox clusters which did not experience the FSGD. This is all the more surprising since in teleosts all six ParaHox genes are distributed across seven instead of four paralogons, and there is not a single complete ParaHox cluster left in the fish lineage [42]. As outlined above, the ParaHox paralogous genomic regions remain identifiable and we wish to emphasize that the paralogous relationship of the RTKs and other genes 3' of the remnants of those ParaHox clusters stay intact. This is because the remaining genes of the ParaHox clusters, and the 3' adjoining RTKs, as well as the genes clock and clock3 that lie directly 3' of the RTKs on the C1 and C2 ParaHox clusters respectively, clearly form paralogous genomic regions.
The C1, C2, D1 and D2 ParaHox paralogons
Using sequence orthology to Astatotilapia burtoni we were able to determine the C and D ParaHox paralogons of Homo sapiens, Mus musculus, Danio rerio, Takifugu rubripes, Tetraodon nigroviridis, Oryzias latipes and Gasterosteus aculeatus (Additional File 1).
We were unable to find the gene clock in the G. aculeatus genome except for two very short blast hits. Since each of the genes of the C1 ParaHox paralogon of this species lies on different contigs within one scaffold, it seems likely that this gene was not correctly assembled in the current release of the stickleback genome. The entire D2 ParaHox paralogon of D. rerio and a major portion of the expected paralogon of G. aculeatus could not be located in the current releases of public genomic databases. The flt4 gene of M. musculus was relocated to another chromosome and the flt4 of H. sapiens was relocated to a location 30 Mb 5' of cdx1. Therefore it was excluded from the following analyses. The cdx1b of Tetraodon nigroviridis is relocated as well and the cdx1b of Oryzias latipes is reversed. All other examined organisms kept the orientations and positions unaltered in reference to the more 5' genes, but the distance to those is always very large (Figure 5).
The orientation and the order of the genes of the C ParaHox paralogons are conserved in all vertebrates species examined (Figures 3 and 4), implying that the orientation and order of these genes have remained unchanged for more than 450 my [53]. The genes gsh2, pdgfrα and kita with its paralogous gene kitb all have a 5' – 3' orientation. The genes kdrb and clock as well as its paralogous gene clock3, show a 3' – 5' orientation (Figures 3 and 4, [42,45]). The genomic architecture of the D1 and D2 ParaHox paralogons is less conserved. The orientation of the genes has stayed the same in all but one species included in this study. Yet, in four genomes the position of a gene compared to pdgfrβa/b and csf1ra/b has changed (Figure 5). Furthermore, the csf1rb gene seems to have been lost in Danio rerio [46]. The gene content of all other paralogons examined in this study was completely conserved.
The presumed ancestral condition of the C ParaHox paralogon that can still be found in mammalian genomes [43,45], it is also conserved in all teleostean a-copies of this ParaHox paralogon (Figures 1a and 3). In all organisms examined here, the b-copy of the C ParaHox complex has lost genes, namely gsh2, pdgfrα and kdrb. The remaining genes of this paralogon nevertheless retained their orientation. A similar scenario can be seen in the D ParaHox paralogon. Here only the b-copy of the gene flt4, a 2R-paralog of kdrb, was lost. We found no trace of clock-like genes 3' of the RTKs so we can not say whether both clock copies were deleted or if the clock precursor was located in the C ParaHox paralogon after the precursor of the C and the D ParaHox paralogons was duplicated. This implies that there never was a clock-like gene in the D1 and D2 ParaHox paralogons.
It seems quite remarkable, that this gene complex maintained both its gene order as well as gene orientation (with the exception of two genes) over very significant evolutionary time spans. Only the lengths of the introns and intergenic regions differ between the species examined. Possible reasons for this conservation might be related to the presence of promoter, enhancer or inhibitor motifs in that complex that influence more than just one gene, so that an inversion or a relocation of one gene would possibly destroy the regulation of the proteins constructed from this and other genes nearby. If such a co-regulation exists, it might explain the maintenance of the gene order and gene orientation over the course of vertebrate evolution. Chiou et al. [54] showed an example of the important role of clustering in the regulation of the expression of biosynthetic genes in A. parasiticus.
Another possibility might be that a regulator is not located immediately next to its corresponding gene but at a distance, and that other genes exist between regulator and corresponding gene. A relocation or inversion in such a case is expected to lead to disruption of the interactions. It has already been shown that regulatory genes or regions lying in a gene cluster are able to control the expression of genes outside of this cluster [55]. Nevertheless, the selective pressures leading to the maintenance of gene clusters are still poorly understood.
In both the C and the D paralogons, only the a-copy retained the ParaHox gene. It was either lost (C2 copy) or relocated (D2 copy, Figure 5). So the b-paralogon in both cases lost more genes than the a-paralogon. Therefore, when comparing the C and the D ParaHox paralogons it is apparent that the a-copies of the ParaHox paralogons C and D of the teleosts are more conserved and show a higher degree of synteny with the mammalian ParaHox paralogons than the b-copies. Interestingly, this finding is similar to the pattern previously found in the Hox clusters [56]. This finding implies that one copy of the paralogon pair evolved faster than the other. That this is always usually the b-copy is explained most easily by the fact that the more conserved (a) copy is much more likely to be discovered and named first.
To further investigate this issue, we performed relative rate tests as described previously [46]. These analyses revealed that the genes of the C paralogons always evolved slower than that of the D paralogons, compared to the human gene (data not shown). Also, we found that the a-paralogon genes always evolved more slowly that the b-copy, except cdx1. cdx1a/b, that are a part of the D1/D2 paralogons respectively, the b-copy evolved more slowly than the a-copy, for unknown reasons – even though the rest of the paralogon follows the normal trend (see Table 1). In this regard, the 3' end and the 5' end of the D1/D2 paralogons seem to experience different evolutionary forces.
Table 1.
Nonparametric Relative Rate Tests of the C and D ParaHox paralogons
| Nucleotide Sequence (first and second codon position) | Amino Acid Sequence | |||||||
| species | para-logons | genea | sites | unique differences | signifi-canceb | sites | unique differences | signifi-canceb |
| T. nigroviridis | Ca-Cb | kita | 1674 | 134 | * | 837 | 43 | * |
| kitb | 176 | 68 | ||||||
| T. rubripes | kita | 1208 | 74 | *** | 603 | 30 | *** | |
| kitb | 140 | 75 | ||||||
| O. latipes | kita | 1524 | 163 | 0.166 | 761 | 63 | 0.391 | |
| kitb | 189 | 73 | ||||||
| G. aculeatus | kita | 1450 | 168 | 0.628 | 724 | 64 | 0.604 | |
| kitb | 177 | 70 | ||||||
| D. rerio | kita | 1864 | 154 | * | 931 | 64 | * | |
| kitb | 192 | 93 | ||||||
| T. nigroviridis | clock | 1574 | 114 | *** | 787 | 55 | *** | |
| clock3 | 217 | 113 | ||||||
| T. rubripes | clock | 1635 | 107 | *** | 816 | 51 | *** | |
| clock3 | 222 | 111 | ||||||
| O. latipes | clock | 1562 | 177 | * | 780 | 112 | ** | |
| clock3 | 143 | 67 | ||||||
| D. rerio | clock | 1592 | 73 | *** | 795 | 32 | *** | |
| clock3 | 203 | 116 | ||||||
| T. nigroviridis | Da-Db | cdx1a | 428 | 121 | *** | 213 | 62 | *** |
| cdx1b | 39 | 15 | ||||||
| T. rubripes | cdx1a | 452 | 116 | *** | 225 | 62 | *** | |
| cdx1b | 55 | 22 | ||||||
| O. latipes | cdx1a | 224 | 30 | * | 111 | 11 | 0.134 | |
| cdx1b | 17 | 5 | ||||||
| T. nigroviridis | pdgfrb1 | 1966 | 222 | *** | 982 | 90 | *** | |
| pdgfrb2 | 328 | 164 | ||||||
| T. rubripes | pdgfrb1 | 2032 | 249 | ** | 1015 | 98 | *** | |
| pdgfrb2 | 323 | 173 | ||||||
| O. latipes | pdgfrb1 | 1999 | 254 | *** | 998 | 97 | *** | |
| pdgfrb2 | 405 | 207 | ||||||
| A. burtoni | pdgfrb1 | 2025 | 216 | *** | 1011 | 72 | *** | |
| pdgfrb2 | 301 | 158 | ||||||
| T. nigroviridis | csf1ra | 1864 | 228 | *** | 904 | 54 | *** | |
| csf1rb | 692 | 323 | ||||||
| T. rubripes | csf1ra | 1842 | 187 | ** | 920 | 59 | *** | |
| csf1rb | 258 | 105 | ||||||
| O. latipes | csf1ra | 1810 | 208 | ** | 904 | 67 | *** | |
| csf1rb | 279 | 119 | ||||||
| A. burtoni | csf1ra | 1856 | 192 | ** | 927 | 62 | * | |
| csf1rb | 247 | 93 | ||||||
| G. aculeatus | csf1ra | 1782 | 183 | *** | 890 | 65 | *** | |
| csf1rb | 306 | 141 | ||||||
a Genes that show statistically significant increase in rate of molecular evolution are indicated in bold.
b χ2 tests: *P < 0.1, **P < 0.01, *** P < 0.001.
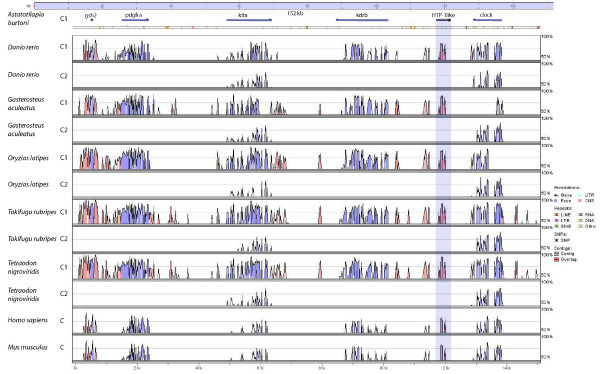
To search for conserved regions that could possibly be promoter regions, enhancers or inhibitors, we performed a mVista plot analysis comparing the C1 ParaHox paralogon of Astatotilapia burtoni with those of Takifugu rubripes, Tetraodon nigroviridis, Danio rerio, Oryzias latipes, Gasterosteus aculeatus, Mus musculus and Homo sapiens. The genes of these paralogons were, as already mentioned, gsh2, pdgfrα, kita, kdrb and clock (Figure 6). The results are similar to our previous analyses of the D Parahox paralogons [46]. Conserved intergenic regions immediately upstream of the genes gsh2, pdgfrα and kita can clearly be detected. Furthermore, there are conserved regions also following the gene clock as well as between it and kdrb. Another conserved region is apparent immediately 3' of kita as well as between the genes gsh2 and pdgfrα (Figure 6). The conserved elements upstream of the genes gsh2 and pdgfrα and a region between kdrb and clock are, at least in part, conserved in all vertebrates examined. Through blasting of a subset of the A. burtoni sequence against the available databases, we found that the conserved sequences between the genes kdrb and clock, are similar to another gene, a transmembrane protein called HTP-1. The other conserved intergenic regions are only conserved in teleost fish. examined and BLAST searches of those regions of the BAC clone 99M12 revealed them to be SINE and LINE elements. As only the fugu specific mask was available, not all elements could be masked efficiently. The only exception to that pattern is the region in Oryzias latipes in the second part of the gene clock. Because of problems in the genome assembly of this organism these data could not be used in the mVista blot analysis and, therefore, medaka had to be omitted from this analysis. As already mentioned, the gene clock of Gasterosteus aculeatus could not be located in its genome assembly. Only two short BLAST hits for clock were found with bl2seq (Figure 6).
Figure 6.
Shuffle-LAGAN vista blot. Comparisons of the C1 and C2 ParaHox paralogons from A. burtoni (query sequence) with T. rubripes, T. nigroviridis, D. rerio, O. latipes, Gasterosteus aculeatus, Mus musculus and H. sapiens. The blue peaks are conserved areas in exons of the genes and in pink conserved intergenic regions are indicated. The genes shown on the graph are from left to right: gsh2, pdgfrα, kita, kdrb and clock.
A comparison of the C1 ParaHox paralogons of Homo sapiens, Astatotilapia burtoni, Danio rerio, Takifugu rubripes, Tetraodon nigroviridis and Gasterosteus aculeatus (Figures 3 and 4) showed that the cichlid sequence is of an intermediate length. It is considerably shorter that H. sapiens (10%),D. rerio (39%) and O. latipes (34%) but longer than T. nigroviridis (150%), T. rubripes (141%) and G. aculeatus (135%) (see Additional file 2). In O. latipes only fragments of clock, the last gene of the paralogon, could be found. Because of seemingly incomplete assembly in this genomic region 34% might not be the final result.
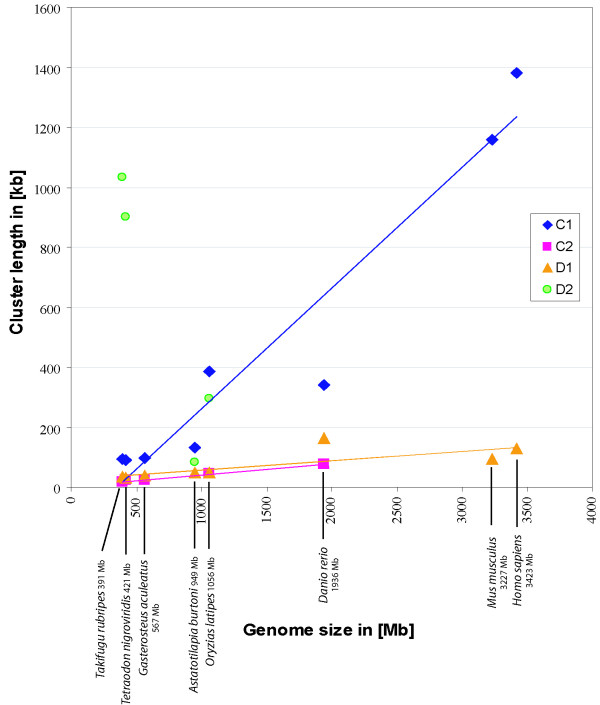
Our previous study [46] showed that the sequence of the cichlid D1 ParaHox paralogon again is of an intermediate length, being shorter than that of D. rerio (29%) but longer that the other D1 paralogons of the other species investigated (O. latipes 102%, T. nigroviridis 148%, T. rubripes 140%, G. aculeatus 125%). In both cases the mammalian sequence is the longest and the sequences of the pufferfishes the shortest (see Additional file 2). To test if this is a paralogon-specific effect or an effect of the different genome sizes of the various organisms, we plotted genome size versus cluster size. Figure 7 shows the comparisons (regression analyses) between the genome sizes of different organisms and the length of the C1, C2, D1 and D2 paralogons. The genome sizes of Homo sapiens, Mus musculus, Danio rerio, Takifugu rubripes, Tetraodon nigroviridis, Gasterosteus aculeatus and Oryzias latipes were taken from the animal genome size database [57]. The genome size estimate of Astatotilapia burtoni was obtained from reference [50].
Figure 7.
Comparison (regression analyses) of the genome size and the size of the C1, C2, D1 and D2 ParaHox paralogons. All genome size estimations, except A. burtoni, were taken from the animal genome size database [57], A. burtoni estimation from [50]. Size estimations of the D1 and D2 clusters were obtained from [46]. The gene flt4 is not included in this analysis. The graph includes the regression lines of the datasets if possible.
While the C1 paralogons of teleosts show a similar genome size to cluster size relationship as the mammalian clusters, the C2 clusters are much more condensed but also show the same trend, namely that the cluster size is linked to overall genome size (Figure 7). The D1 paralogons are also much more condensed than the C1 paralogons, but they also display a linear relationship between genome and cluster size, including also the mammalian sequences. An obvious deviation from previously described pattern can be seen in the D2 paralogons of the pufferfishes. Relative to their very compact genome size, the D2 paralogon is surprisingly large. A possible reason for this could be that the maximal condensation of this cluster has already been reached and a further condensation might be detrimental in terms of selection. We can only speculate on this, but the minimum absolute size of the ParaHox paralogons might be determined by the necessary spatial relationships of the individual transcriptional units within these gene complexes and regulatory regions might need to be maintained at a minimal distance in the intergenic regions of adjacent genes in order to maintain the proper function of these genes. The Astatotilapia burtoni D2 ParaHox paralogon could not be included in this comparison because the gene cdx1b is not on the investigated BAC clone.
Gene cluster breakup and gene retention after genome duplications
The FSGD provides the opportunity to study genomes following a whole genome duplication event [[33] and references therein]. For the Hox gene clusters of teleosts, it has been observed before [[39] and references therein] that, although all fish genomes studied so far vary in the gene content and even number of Hox gene clusters, the total number of Hox genes contained in their genomes is about the same as in the genomes of tetrapods, which did not experience this WGD. It has been suggested that particularly the Hox gene clusters are, typically, maintained more or less intact, because they are likely to be strongly regulated by sequential activation and cluster completeness is necessitated by corrected interdigitated gene control [58].
What seems remarkable as well is that the evolutionary forces keeping Hox gene number rather constant seem to be stronger than those that maintain the cohesion and physical linkage on chromosomes of individual clusters following a WGD. Mulley et al. [42] noted that the ParaHox cluster stayed intact in ancestral fish lineages such as Amia and Polypterus, yet noted the fragmentation of the ParaHox clusters in teleosts, that happened due to gene loss and not because of transpositions or inversions [42]. The FSGD duplicated all genomic regions including the clustered sets of homeobox genes such as Hox, ParaHox and NK. The selection pressures that maintained those clusters intact in part of the metazoans, seem to be relaxed, as for many of these gene clusters, several genes seem to have been lost [59], despite the fact that these, often apparently co-regulated arrays of genes, seem to share enhancers and are regulated in an interdigitated fashion (Figure 1). Mulley et al. [42] proposed that the maintenance of a gene cluster is based on interdigitated and/or shared enhancers. The FSGD duplicated not only the genes but also the enhancers and therefore might have released the need for a tight clustering. Our analysis of the ParaHox clusters in teleosts supports this idea in so far as the ParaHox clusters are broken up. Yet, the total number of six ParaHox genes is maintained in post duplication teleost genomes. If the comparison is extended to a larger paralogon than the set of ParaHox genes alone – as was done in this study -it becomes clear that in larger genomic regions there the constancy of gene numbers does not persist. Our analysis shows that some, although not all, additional duplicated genes flanking the ParaHox clusters were retained following a WGD (Figure 1). This might imply that different selective forces such as increased tolerance to more gene product, due to the doubled number of genes, or functional changes (sub-, neofunctionalization) of those genes might be acting. This finding might argue that although differential gene loss on different chromosomal regions is permitted following a WGD through genetic redundancy of cis-regulatory elements, the overall constancy of gene number is strongly selected for by balancing selection at least for transcription factors such as ParaHox genes. Balancing selection might be acting on trans-regulatory mechanisms to countact possibly negative effects of dosage differences. Moreover, possibly weaker selective forces against duplicate genes might permit the retention of probably not co-regulated genes outside of gene clusters after a WGD on one hand. It seems plausible that these different selective forces might also have to do with not only their arrangements in clusters, but also which kind of gene is duplicated (e.g., regulatory genes vs. housekeeping genes). Again, selection might act more strongly in bringing about the loss of interdigitated genes within cluster following a WGD to maintain a modal gene number per genome of these clustered homeobox genes in order to reduce potentially negative changes in dosage following a WGD. The fact, that the number of ParaHox genes before and after the FSGD remained unchanged, indicates a possibly strong regulatory gene dose restriction that would select for the rapid loss of "superfluous" genes. With the exception of the gene cdx1 no gene of the ancestral ParaHox cluster was retained in two copies. Possibly one of the two cdx1 genes may be compensating for the loss of cdx2 gene, hence the retention of two cdx1 genes (Figure 1b, [42]).
Recently Negre and Ruiz [60] have discovered a surprising diversity of Hox gene cluster architectures in different species of Drosophila. Since breaks and inversions were found not too infrequently, they argue that not the integrity and organization of Hox clusters is the strongest target of selection. Rather they argue that functional constraints on individual Hox genes might be acting more forcefully on genomes so that functional sets of homeobox genes are maintained in the genome, which are not necessarily physically linked with unbroken colinearity. Other studies showed that an intact cluster is only important for temporal and not for special colinearity. In Drosophila where development is so rapid that almost all the Hox genes are activated at the same time, the cluster is permitted to be interrupted [[61] and references therein]. Similar reasoning might explain the sitution we describe for "dissolved" ParaHox parologons. Their genomically fixed gene content and orientation in teleost genomes, but their dispersed distribution over seven instead of four chromosomal regions would support the hypothesis that overall gene content is more strongly selected for than the integrity of gene clusters.
Conclusion
We demonstrated the orthologous relationship of the genes of the C and D ParaHox paralogons (Figure 2). Relative rate tests revealed that with the exception of one gene the a-copy always evolves more slowly than the b-copy, the exception being the ParaHox gene cdx1, where the b-copy evolves significantly slower. The relative rate tests also show that the C paralogons evolve more slowly than the D paralogons.
A mVista analysis of the D clusters was performed in an earlier study [46]. We found a number of conserved genomic regions in the C1 ParaHox paralogon that were located in intergenic regions. One conserved sequence block, located at the position 119–130 kb on the A. burtoni BAC clone 99M12, was confirmed to be another gene, the transmembrane protein HPT-1, by BLAST search. We also found evidence that the ParaHox paralogon of the pufferfishes is apparently close to the maximal possible reduction in size.
Despite having undergone an additional genome duplication the total number of ParaHox genes in the genome of teleost fish is maintained at six genes that are distributed over seven chromosomal regions instead of four as in the genomes of tetrapods. Other genes that are physically linked with the ParaHox genes in the same paralogon were also reduced in number following the FSGD. However, while typically ten of these are found in tetrapods 14 are maintained in teleost fish genomes. We discuss possible selective reasons for keeping modal numbers of homoebox genes constant throughout hundreds of millions of years of evolution while permitting to differentially loose ParaHox genes on some ParaHox paralogons.
Future research should include the description of possible binding sites in the conserved elements and functional studies of those putative regulatory elements found by in silico analyses.
Methods
BAC Library screening & Shotgun Sequencing
We previously constructed a BAC library of the East African haplochromine cichlid fish Astatotilapia burtoni [50]. This library was screened for kita positive clones with the kita specific primer set Burt-Kit-F-474/Burt-Kit_R-672 according to [46]. Using universal primers (gsh2_Ex1_For (AGAYCCCAGRAGATACCACT) and gsh2_Ex2.3_R (GTGCGCGCTCCTCTGGGTG)) designed on known teleost sequences, we confirmed the presence of the gsh2 gene on the BAC clone. The BAC plasmids of the recovered clones were extracted using the Large-Construct Kit (Qiagen) according to the manufacturer's manual and then sheared by sonification. The fraction of 2–3 kb was recovered from an agarose gel and blunt-end-ligated into the pUC18 vector of Roche and later electro-transformed into "Electro Max DH10B T1 Phage Resistant Cells" (Invitrogen). The subclones were grown in standard LB-medium (0.5 mg/ml ampicillin). The plasmid DNA was recovered using standard methods. The clones were sequenced directly using a standard M13F/M13R primer set on an ABI3100 automatic DNA sequencer (Applied Biosystems).
Contig Assembly
The obtained sequences were quality trimmed by hand and checked for vector sequences using Sequencher 4.2 (Gene Codes Corporation). The same software was used for the contig assembly at the setting "dirty data", with a sequence similarity of 85% and an overlap of 20 bp. The full sequence of the genome of E. coli from the GENBANK database [52] was added to the analyses, so that all E. coli contaminated reads were filtered out of the assembly. Gaps between contigs were closed with gap spanning primers, designed with Primer3 [62]. For further analyses the remaining gaps were closed by 33 N's each.
The contigs of the BAC clone 99M12 were checked for corresponding forward/reverse clones in other contigs and a contig map was drawn. To check this map, the contigs were assembled into one single sequence according to the contig map. Using the tool bl2seq (align two sequences) (GENBANK database) [52], the contigs were BLAST-searched against chromosome 11 of Tetraodon nigroviridis containing its C1 ParaHox cluster. The contig map was then corrected using the information from the bl2seq analysis.
Sequence Annotation
The ontology of the genes sequenced were determined by sequence comparisons with the available genomes of Takifugu rubripes [63], (version 4.0), Tetraodon nigroviridis [64], (version 1–64), Oryzias latipes [65], (version 200506), Gasterosteus aculeatus [66], (version 41) and already annotated genes from Danio rerio, Mus musculus and Homo sapiens were taken from GENBANK [52]. The provided annotations of the Homo sapiens, Mus musculus and Danio rerio sequences from GENBANK database [52] were used to help to identify the intron/exon structure of the respective genes in Takifugu rubripes, Tetraodon nigroviridis and Oryzias latipes. In some cases the Danio rerio sequence could not be included into the analyses due to apparent miss-assemblies.
Phylogenetic and Sequence Analyses
For phylogenetic analyses, Nexus files were processed via PAUP* [67] to eliminate positions that could not be aligned. The appropriate models of molecular evolution were estimated using the program modelgenerator [68]. Maximum likelihood trees and bootstrapping (1000 replicates) were calculated in PHYML [69]. Bayesian Inference was performed in Mr. Bayes 3.1 [70,71] (1,000,000 generations/5000 burnin).
Vista Plots were obtained via the mVista option on the Vista homepage [72]. The alignment program used was LAGAN (Global multiple alignment of finished sequences) [73]. For Homo sapiens the human/primate-specific RepeatMasker and for Mus musculus the mouse/rat/rodent specific RepeatMasker was used. For all other sequences the fugu-specific RepeatMasker was used as a stand in.
Authors' contributions
This study was conceived by SH, WS, IB and AM. Most of the laboratory work was done by NS and IB. In silico analyses were conducted by NS with the help of SH, WS and IB. The manuscript was drafted by NS and read and revised by all authors.
Supplementary Material
Location of the genes of the C1 and C2 ParaHox paralogon. List of the genes used for the computation of the trees via PHYML and Mr. Bayes analyses, including the orientation of the gene (→ means 5'-3'; ← means 3'-5' orientation) and the database the DNA sequence was taken from. All sequences from NCBI (National Center for Biotechnology Information) [52] were taken as annotated there; the sequences from the other databases (MGP (Medaka Genome Project) [65], version 200506) and Ensembl [66]T. nigroviridis version Tetraodon7, T. rubripes version Fugu4.0, O. latipes version Medaka1, G. aculeatus version BroadS1,D. rerio version Zv6) were annotated by hand.
Comparison of the genome size and the size of the C1, C2, D1 and D2 ParaHox paralogons. All genome size estimations, except A. burtoni from the animal genome size database [57], A. burtoni estimation from (Lang et al. 2006); size estimation of the D1 and D2 cluster from (Braasch et al. 2006). The gene flt4 is not included in this analysis.
Acknowledgments
Acknowledgements
Support from the Deutsche Forschungsgemeinschaft (DFG), from the European Community, and the Landesstiftung Baden-Württemberg GmbH is gratefully acknowledged.
Contributor Information
Nicol Siegel, Email: Nicol.Siegel@meduniwien.ac.at.
Simone Hoegg, Email: Simone.Isabel.Hoegg@uni-konstanz.de.
Walter Salzburger, Email: Walter.Salzburger@unil.ch.
Ingo Braasch, Email: ingo.braasch@biozentrum.uni-wuerzburg.de.
Axel Meyer, Email: Axel.Meyer@uni-konstanz.de.
References
- Nelson JS. Fishes of the world. Hoboken, New Jersey , John Wiley & Sons; 2006. [Google Scholar]
- Fryer G, Iles TD. The Cichlid Fishes of the Great Lakes of Africa: Their Biology and Evolution. Oliver and Boyd, Edinburgh. 1972.
- Kocher TD. Adaptive evolution and explosive speciation: the cichlid fish model. Nature Reviews Genetics. 2004;5:288–298. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- Kornfield I, Smith PF. African Cichlid Fishes: Model Systems for Evolutionary Biology. Annual Reviews in Ecology and Systematics. 2000;31:163–196. doi: 10.1146/annurev.ecolsys.31.1.163. [DOI] [Google Scholar]
- Meyer A. Phylogenetic relationships and evolutionary processes in East African cichlids. Trends in Ecology and Evolution. 1993;8:279–284. doi: 10.1016/0169-5347(93)90255-N. [DOI] [PubMed] [Google Scholar]
- Salzburger W, Meyer A. The species flocks of East African cichlid fishes: recent advances in molecular phylogenetics and population genetics. Naturwissenschaften. 2004;91:277–290. doi: 10.1007/s00114-004-0528-6. [DOI] [PubMed] [Google Scholar]
- Schluter D. The ecology of adaptive radiation. Oxford University Press, New York. 2000.
- Stiassny MLJ, Meyer A. Cichlids of the Rift Lakes. Scientific American. 1999;February:64–69. [Google Scholar]
- Turner GF, Burrows MT. A model of sympatric speciation by sexual selection. Proceedings Biological sciences / The Royal Society B. 1995;260:287–292. doi: 10.1098/rspb.1995.0093. [DOI] [Google Scholar]
- Barlow GW. The cichlid fishes. Nature's grand experiment in evolution. Cambridge , Perseus Publishing; 2000. [Google Scholar]
- Salzburger W, Niederstatter H, Brandstatter A, Berger B, Parson W, Snoeks J, Sturmbauer C. Colour-assortative mating among populations of Tropheus moorii, a cichlid fish from Lake Tanganyika, East Africa. Proc Biol Sci. 2006;273:257–266. doi: 10.1098/rspb.2005.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem KF. Adaptive significance of intra- and interspecific differences in the feeding repertoires of cichlid fishes. American Zoologist. 1980;20:295–314. [Google Scholar]
- Garcia-Fernandez J. Hox, ParaHox, ProtoHox: facts and guesses. Heredity. 2005;94:145–152. doi: 10.1038/sj.hdy.6800621. [DOI] [PubMed] [Google Scholar]
- Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994:125–133. [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. Springer-Verlag, New York. 1970.
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999;11:699–704. doi: 10.1016/S0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Durand D, Hoberman R. Diagnosing duplications--can it be done? Trends in genetics. 2006;22:156–164. doi: 10.1016/j.tig.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Long M. Evolution of novel genes. Curr Opin Genet Dev. 2001;11:673–680. doi: 10.1016/S0959-437X(00)00252-5. [DOI] [PubMed] [Google Scholar]
- Tymowska J, Fischberg M, Tinsley RC. The karyotype of the tetraploid species Xenopus vestitus Laurent (Anura: pipidae) Cytogenetics and cell genetics. 1977;19:344–354. doi: 10.1159/000130828. [DOI] [PubMed] [Google Scholar]
- Beetschen JC. [5 generations of polyploid individuals in the salamander, Pleurodeles watlii Michah] C R Seances Soc Biol Fil. 1967;161:930–936. [PubMed] [Google Scholar]
- Danzmann RG, Cairney M, Davidson WS, Ferguson MM, Gharbi K, Guyomard R, Holm LE, Leder E, Okamoto N, Ozaki A, Rexroad CE, 3rd, Sakamoto T, Taggart JB, Woram RA. A comparative analysis of the rainbow trout genome with 2 other species of fish (Arctic charr and Atlantic salmon) within the tetraploid derivative Salmonidae family (subfamily: Salmoninae) Genome. 2005;48:1037–1051. doi: 10.1139/g05-067. [DOI] [PubMed] [Google Scholar]
- Leggatt RA, Iwama GK. Occurrence of polyploidy in the fishes. Reviews in Fish Biology and Fisheries. 2004;13:237–246. [Google Scholar]
- Hanson RE, Islam-Faridi MN, Percival EA, Crane CF, Ji Y, McKnight TD, Stelly DM, Price HJ. Distribution of 5S and 18S-28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma. 1996;105:55–61. doi: 10.1007/BF02510039. [DOI] [PubMed] [Google Scholar]
- Islam N, Tsujimoto H, Hirano H. Proteome analysis of diploid, tetraploid and hexaploid wheat: towards understanding genome interaction in protein expression. Proteomics. 2003;3:549–557. doi: 10.1002/pmic.200390068. [DOI] [PubMed] [Google Scholar]
- Patterson JT, Larson SR, Johnson PG. Genome relationships in polyploid Poa pratensis and other Poa species inferred from phylogenetic analysis of nuclear and chloroplast DNA sequences. Genome. 2005;48:76–87. doi: 10.1139/g04-102. [DOI] [PubMed] [Google Scholar]
- Vandepoele K, De Vos W, Taylor JS, Meyer A, Van de Peer Y. Major events in the genome evolution of vertebrates: Paranome age and size differs considerably between ray-finned fishes and land vertebrates. Proc Natl Acad Sci USA. 2004;101:1638–1643. doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Suzuki T, Yan YL, Pomeroy J, Singer A, Amemiya C, Postlethwait JH. Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish. Genome Res. 2004;14:1–10. doi: 10.1101/gr.1717804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegg S, Brinkmann H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. Journal of molecular evolution. 2004;59:190–203. doi: 10.1007/s00239-004-2613-z. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Van de Peer Y, Braasch I, Meyer A. Comparative genomics provides evidence for an ancient genome duplication event in fish. Phil Trans R Soc Lond B Biol Sci. 2001;356:1661–1679. doi: 10.1098/rstb.2001.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Gu X, Wang Y, Gu J. Age distribution of human gene families shows significant roles of both large- and small-scale duplications in vertebrate evolution. Nature Genetics. 2002;31:205–209. doi: 10.1038/ng902. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Wittbrodt J, Meyer A, Schartl M. More genes in fish? BioEssays. 1998;20:511–515. doi: 10.1002/(SICI)1521-1878(199806)20:6<511::AID-BIES10>3.0.CO;2-3. [DOI] [Google Scholar]
- Christoffels A, Koh EG, Chia JM, Brenner S, Aparicio S, Venkatesh B. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol Biol Evol. 2004;21:1146–1151. doi: 10.1093/molbev/msh114. [DOI] [PubMed] [Google Scholar]
- Hoegg S, Meyer A. Hox clusters as models for vertebrate genome evolution. Trends in genetics. 2005;21:421–424. doi: 10.1016/j.tig.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Garcia-Fernandez J, Holland PW. Archetypal organization of the amphioxus Hox gene cluster. Nature. 1994;370:563–566. doi: 10.1038/370563a0. [DOI] [PubMed] [Google Scholar]
- Chourrout D, Delsuc F, Chourrout P, Edvardsen RB, Rentzsch F, Renfer E, Jensen MF, Zhu B, de Jong P, Steele RE, Technau U. Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature. 2006;442:684–687. doi: 10.1038/nature04863. [DOI] [PubMed] [Google Scholar]
- Mulley JF, Chiu CH, Holland PW. Breakup of a homeobox cluster after genome duplication in teleosts. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10369–10372. doi: 10.1073/pnas.0600341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrier DE, Dewar K, Cook A, Chang JL, Hill-Force A, Amemiya C. The chordate ParaHox cluster. Curr Biol. 2005;15:R820–2. doi: 10.1016/j.cub.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Minguillon C, Garcia-Fernandez J. Genesis and evolution of the Evx and Mox genes and the extended Hox and ParaHox gene clusters. Genome Biol. 2003;4:R12. doi: 10.1186/gb-2003-4-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prohaska SJ, Stadler PF. Evolution of the vertebrate parahox clusters. J Exp Zoolog B Mol Dev Evol. 2006 doi: 10.1002/jez.b.21099. [DOI] [PubMed] [Google Scholar]
- Braasch I, Salzburger W, Meyer A. Asymmetric evolution in two fish-specifically duplicated receptor tyrosine kinase paralogons involved in teleost coloration. Mol Biol Evol. 2006;23:1192–1202. doi: 10.1093/molbev/msk003. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Turner JM. Temporal and cellular requirements for Fms signaling during zebrafish adult pigment pattern development. Development. 2003;130:817–833. doi: 10.1242/dev.00307. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Ransom DG, Paw B, Zon LI, Johnson SL. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development. 2000;127:3031–3044. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Rawls JF, Pratt SJ, Whitfield TT, Johnson SL. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development. 1999;126:3425–3436. doi: 10.1242/dev.126.15.3425. [DOI] [PubMed] [Google Scholar]
- Lang M, Miyake T, Braasch I, Tinnemore D, Siegel N, Salzburger W, Amemiya CT, Meyer A. A BAC library of the East African haplochromine cichlid fish Astatotilapia burtoni. J Exp Zoolog B Mol Dev Evol. 2006;306:35–44. doi: 10.1002/jez.b.21068. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov
- Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- Chiou CH, Miller M, Wilson DL, Trail F, Linz JE. Chromosomal location plays a role in regulation of aflatoxin gene expression in Aspergillus parasiticus. Appl Environ Microbiol. 2002;68:306–315. doi: 10.1128/AEM.68.1.306-315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MS, Yu J, Nierman WC, Kim HS, Pritchard B, Jacobus CA, Bhatnagar D, Cleveland TE, Payne GA. The aflatoxin pathway regulator AflR induces gene transcription inside and outside of the aflatoxin biosynthetic cluster. FEMS Microbiol Lett. 2006;255:275–279. doi: 10.1111/j.1574-6968.2005.00084.x. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Takahashi K, Lynch V, Prohaska SJ, Fried C, Stadler PF, Amemiya C. Molecular evolution of duplicated ray finned fish HoxA clusters: increased synonymous substitution rate and asymmetrical co-divergence of coding and non-coding sequences. J Mol Evol. 2005;60:665–676. doi: 10.1007/s00239-004-0252-z. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Animal Genome Size Database. http://www.genomesize.com. 2005.
- Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Garcia-Fernandez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005;6:881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- Negre B, Casillas S, Suzanne M, Sanchez-Herrero E, Akam M, Nefedov M, Barbadilla A, de Jong P, Ruiz A. Conservation of regulatory sequences and gene expression patterns in the disintegrating Drosophila Hox gene complex. Genome Res. 2005;15:692–700. doi: 10.1101/gr.3468605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro AS, Ferrier DE. Hox genes are not always Colinear. Int J Biol Sci. 2006;2:95–103. doi: 10.7150/ijbs.2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Bioinformatics Methods and Protocols: Methods in Molecular Biology Humana Press Totowa NJ. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- JGI Database http://www.jgi.doe.gov/
- Genoscope http://www.genoscope.cns.fr/externe/tetranew/
- Medaka Genome Project http://dolphin.lab.nig.ac.jp/medaka/
- Ensembl Genome Browser http://www.ensembl.org/index.html
- Rogers JS, Swofford DL. A fast method for approximating maximum likelihoods of phylogenetic trees from nucleotide sequences. Systematic biology. 1998;47:77–89. doi: 10.1080/106351598261049. [DOI] [PubMed] [Google Scholar]
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McLnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evolutionary Biology. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA : visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome research. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Location of the genes of the C1 and C2 ParaHox paralogon. List of the genes used for the computation of the trees via PHYML and Mr. Bayes analyses, including the orientation of the gene (→ means 5'-3'; ← means 3'-5' orientation) and the database the DNA sequence was taken from. All sequences from NCBI (National Center for Biotechnology Information) [52] were taken as annotated there; the sequences from the other databases (MGP (Medaka Genome Project) [65], version 200506) and Ensembl [66]T. nigroviridis version Tetraodon7, T. rubripes version Fugu4.0, O. latipes version Medaka1, G. aculeatus version BroadS1,D. rerio version Zv6) were annotated by hand.
Comparison of the genome size and the size of the C1, C2, D1 and D2 ParaHox paralogons. All genome size estimations, except A. burtoni from the animal genome size database [57], A. burtoni estimation from (Lang et al. 2006); size estimation of the D1 and D2 cluster from (Braasch et al. 2006). The gene flt4 is not included in this analysis.