Abstract
High mobility group (HMG) A1 proteins are subject to a number of post-translational modifications, which may regulate their function in gene transcription and other cellular processes. We examined, by using mass spectrometry, the acetylation of HMGA1a and HMGA1b proteins induced by histone acetyltransferases p300 and PCAF in vitro and in PC-3 human prostate cancer cells in vivo. It turned out that five lysine residues in HMGA1a, i.e., Lys-14, Lys-64, Lys-66, Lys-70, and Lys-73, could be acetylated by both p300 and PCAF. We further quantified the level of acetylation by analyzing, with LC-MS/MS, the proteolytic peptides of the in-vitro or in-vivo acetylated HMGA1 proteins where the unmodified lysine residues were chemically derivatized with a perdeuterated acetyl group. Quantification results revealed that p300 and PCAF exhibited different site preferences for the acetylation; the preference of p300 acetylation followed the order of Lys-64~Lys-70 > Lys-66 > Lys-14~Lys73, whereas the selectivity of PCAF acetylation followed the sequence of Lys-70~Lys-73 > Lys-64~Lys-66 > Lys-14. HMGA1b was acetylated in a very similar fashion as HMGA1a. We also demonstrated that C-terminal phosphorylation of HMGA1 proteins did not affect the in-vitro acetylation of the two proteins by either p300 or PCAF. Moreover, we examined the acetylation of lysine residues in HMGA1a and HMGA1b isolated from PC-3 human prostate cancer cells. Our results showed that all the above five lysine residues were also acetylated in vivo, with Lys-64, Lys-66 and Lys-70 in HMGA1a exhibiting higher levels of acetylation than Lys-14 and Lys-73.
Introduction
High mobility group (HMG) proteins, comprising three families of structurally unrelated proteins including HMGA, HMGB, and HMGN, are non-histone chromosomal proteins that are thought to play important roles in the assembly of chromatin and in the regulation of transcription in higher eukaryotic cells [1]. HMGA1 proteins contain three independent DNA-binding regions, called AT-hook motifs, which bind to the minor groove of AT-rich DNA sequences both in vitro and in vivo [1–3]. HMGA1a and HMGA1b, formerly known as HMG-I and HMG-Y, respectively, are translated from the splicing variants of a single gene (HMGA1) and they are identical in sequence except for an 11-amino acid internal deletion in HMGA1b (Figure 1) [4, 5]. Both HMGA1a and HMGA1b are recognized as architectural transcription factors that up- or down-regulate the transcriptional activity of a number of human genes [6, 7]. Despite the structural similarities between HMGA1a and HMGA1b, these two proteins may have distinct biological functions [8–10].
Figure 1.

The sequences of HMGA1a and HMGA1b. The acetylated lysine residues identified in this study were highlighted in bold, and peptides containing these lysine residues were underlined.
Among the highly modified nuclear proteins, HMGA1 proteins adopt a number of post-translational modifications (PTMs) including acetylation, methylation and phosphorylation. Phosphorylation of HMGA1 proteins was first detected by Lund et al. [11] in Ehrilich ascites cells. Further research revealed that HMGA1 proteins were phosphorylated by several kinases such as cyclin-dependent kinase 1 (previously known as cdc2 kinase) [12, 13], protein kinase C (PKC) [14] and protein kinase CK2 [15]. These phosphorylations attenuated the DNA binding affinities of mammalian HMGA1a [14, 16, 17] and HMGA1b [17, 18].
In addition to phosphorylation, methylation was also detected in HMGA1 proteins [8, 19–23]. Dimethylation of both arginine and lysine residues in HMGA1a was reported and it might be correlated with the metastatic potential and apoptosis of cells [19–21]. Recently, we reported that Arg-25 in HMGA1a, but not in HMGA1b, was both mono- and dimethylated in PC-3 human prostate cancer cells, with the dimethylation being in either symmetric or asymmetric form [24]. Importantly, some of the above phosphorylation and methylation were also found in HMGA1a and HMGA1b proteins isolated from human breast cancer tissues [25].
Other than phosphorylation and methylation, acetylation of HMGA1 proteins was also detected [8, 9, 19, 26–28]. In this respect, Lys-64 and Lys-70 in HMGA1a protein can be acetylated by CBP (CREB-binding protein) and PCAF (p300/CBP-associated factor), respectively [26, 27]. In addition, the acetylation at these two sites conferred distinct biological outcomes; the acetylation of Lys-64 destabilized the enhanceosome, whereas the acetylation of Lys-70 potentiated transcription of interferon-β gene by stabilizing the enhanceosome and preventing the acetylation of HMGA1a by CBP [26, 27]. Moreover, multiple acetylation sites in HMGA1 proteins from MCF-7 human breast cancer cells have been suggested, though the assignment of these modifications to specific lysine residues was not made [8]. Very recently, we reported that Lys-14 in HMGA1a and HMGA1b was acetylated in PC-3 cells. We, however, were not able to identify any acetylated peptides containing Lys-64 and/or Lys-70 from the tryptic digestion mixture of HMGA1a [28].
Mass spectrometry is commonly used for the identification of PTMs in proteins [29, 30]. However, PTMs often alter the ionization efficiency of the peptide being modified and, as a consequence, the observed relative abundances of ions corresponding to the modified and unmodified forms of a peptide may not reflect accurately the level of modification. This problem can be resolved by using stable isotope-labeled proteins or peptides, which are chemically identical to the naturally occurring proteins or peptides [31–33]. Incorporation of non-radioactive isotopes into peptides or proteins of interest can be achieved either in cell culture [34, 35] or after the extraction of peptides/proteins from cells or tissues [36, 37]. In this regard, quantitative analysis of phosphorylation [38, 39], glycosylation [40, 41] and acetylation [42] has been reported.
Herein, we adopted the quantification method introduced by Smith et al. [42], which was developed to quantify the levels of acetylation at individual lysine residues in the N-terminal tail of histone H4, and examined systematically the sites and levels of acetylation of HMGA1 proteins induced by histone acetyltransferases p300 and PCAF in vitro. Because HMGA1 proteins isolated from cancer cells showed simultaneous acetylation and phosphorylation [28, 43], we further assessed how C-terminal constitutive phosphorylation affects the acetylation of the two proteins. Moreover, we investigated the acetylation of lysine residues in HMGA1a and HMGA1b isolated from PC-3 human prostate cancer cells.
Experimental
Purification of HMGA1 proteins
Full-length recombinant human HMGA1a and HMGA1b proteins were overexpressed in E. coli BL21 DE3 pLysS cells (Invitrogen, Carlsbad, CA) followed by extraction with 5% perchloric acid (PCA) as reported previously [44, 45]. Recombinant HMGA1 proteins were further purified on an Agilent 1100 system (Agilent Technologies, Palo Alto, CA) by using a 4.6 × 250 mm C4 column (Grace Vydac, Hesperia, CA). The flow rate was 1.0 mL/min, and a 60-min gradient of 5–50% CH3CN in 0.1% aqueous solution of trifluoroacetic acid (TFA) was employed. The purified proteins were then quantified by Bradford protein assay (Bio-Rad, Hercules, CA).
The HMGA1 proteins were also isolated from PC-3 human prostate cancer cells following previously described procedures [24]. Briefly, the PC-3 cells were cultured in F-12 media (ATCC, Manassas, VA) supplemented with 10% fetal bovine serum (Invitrogen) and 5% CO2 at 37°C. Cells were harvested and homogenized by sonication in the lysis buffer followed by PCA extraction. The HMGA1 proteins were then isolated from the PCA-soluble fractions by using HPLC and a 4.6 × 250 mm C4 column (Varian, Walnut Creek, CA).
In-vitro phosphorylation of HMGA1 proteins by protein kinase CK2
Recombinant HMGA1a or HMGA1b (15 μg) was incubated with 600 U of protein kinase CK2 (New England Biolabs, Beverly, MA) and 200 μM ATP at 37°C for 1 h in a 150-μL reaction buffer supplied by the vendor. The phosphorylated HMGA1 proteins were then isolated from the reaction mixture by using HPLC on the Agilent 1100 system with a 2.0×250 mm C4 column (Phenomenex, Torrance, CA). The flow rate was 200 μL/min, and a 40-min gradient of 5–40% CH3CN in 0.1% aqueous solution of TFA was employed. The chromatogram was obtained by absorbance detection at 220 nm. Fractions containing phosphorylated HMGA1a or HMGA1b were collected and subjected to MALDI-MS analysis to confirm the phosphorylated products.
In-vitro acetylation of HMGA1 proteins by p300 and PCAF
Recombinant and CK2-phosphorylated HMGA1 proteins were acetylated by the HAT (histone acetyltransferase) domain of p300 (Upstate, Temecula, CA) or PCAF (Upstate) at enzyme-to-substrate molar ratios of 1:20 and 1:30, respectively. The acetylation reactions were carried out in the presence of 1.5 nmol acetyl CoA in a HAT buffer (50 mM Tris-HCl, pH 8.0, 0.1 mM EDTA, 1.0 mM dithiothreitol and 10% glycerol) at 30°C for 1 h [46, 47].
Isotope labeling and enzymatic digestion
Chemical derivatization of unmodified lysine residues in HMGA1a or HMGA1b was carried out by incubating the protein with 50 μL deuterated acetic acid and 10 μL deuterated acetic anhydride at room temperature for 14 hrs. Deuterated proteins were then dried in a Speedvac concentrator and resuspended in 50 mM NH4HCO3 followed by enzymatic digestion with sequencing-grade modified trypsin or Glu-C (Roche Applied Science, Indianapolis, IN) at respective enzyme-to-substrate ratios of approximately 1:25 and 1:10 at 37°C overnight.
Mass spectrometry
Matrix-assisted laser desorption/ionization (MALDI)-MS measurements were performed on a QSTAR XL quadrupole/time-of-flight (TOF) mass spectrometer equipped with an o-MALDI ion source (Applied Biosystems, Foster city, CA). Purified protein samples were dissolved in an aqueous solution of 0.1% TFA, and sample aliquots (0.5 μL) were mixed with an equal volume of α-cyano-4-hydroxy-cinnamic acid (CHCA) saturated in 50% CH3CN containing 0.1% TFA. The mass accuracy in MS/MS mode was approximately 10–30 ppm with external calibration. The digested peptides were desalted with C18 ZipTip (Millipore, Billerica, MA) and mixed with an equal volume of CHCA matrix solution before subjecting to mass spectrometric analysis.
Most LC-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) experiments were carried out by coupling directly the effluent from a Zorbax SB-C18 capillary column (0.5×150 mm, 5 μm in particle size, Agilent Technologies) to a Global Ultima Q-TOF mass spectrometer (Micromass, UK). Sequential elution of peptides was accomplished by using a 65-min linear gradient of 2–50% acetonitrile in 0.1% formic acid delivered by the Agilent 1100 capillary HPLC pump at a flow rate of 6 μL/min. The spray voltage was 3.32 V, and the desolvation and source-block temperature were maintained at 120°C and 80°C, respectively. Nitrogen was used as the cone and desolvation gas, while helium was used as the collision gas.
Some LC-ESI-MS/MS experiments were also performed on an LTQ linear ion trap mass spectrometer (Thermo Electron Co., San Jose, CA). MS/MS experiments were carried out in either the data-dependent scan mode or the pre-selected ion mode with a relative collision energy of 35%. The spray voltage was 4.5 kV, and the temperature for the heated capillary was 275°C.
Quantitative assessment of the levels of acetylation was based on LC-MS/MS results acquired on the Q-TOF mass spectrometer, which was set up to monitor specifically the fragmentations of the protonated ions of the three peptides housing the acetylated lysine residues. The quadruple mass filter before the TOF analyzer was set with an LM and HM resolution of 2.0 (arbitrary units), which was equivalent to a 10–15 m/z window for the transmission of precursor ions. The large isolation width was chosen because it allowed for the precursor ions of both the acetylated (with an H3-Ac group) and the unacetylated forms (with a D3-Ac group introduced by chemical derivatization, vide supra) of a peptide to be selected for fragmentation. A potential drawback of using a large isolation width lies in that it may also allow for other ions with m/z values lying in the chosen m/z window to be selected for fragmentation and complicate the product-ion spectra. Nevertheless, under these experimental conditions, almost all ions found in the resulting MS/MS could be attributed to form from the cleavages of the peptide under investigation (vide infra).
Relative acetylation levels of lysine residues were quantified by calculating the ratio of peak intensities for the H3-Ac- and D3-Ac-labeled precursor or product ions. In this respect, five to fourteen scans were averaged to give the spectrum used for the quantitative assessment of the level of acetylation.
Results
In-vitro acetylation of recombinant HMGA1a and HMGA1b
CBP and its closely related p300 were discovered as transcriptional co-activators for a number of sequence-specific factors that integrate diverse signaling pathways [48]. These two proteins and their associated factor PCAF have intrinsic histone acetyltransferase activity [49, 50]. They can induce the acetylation of lysine residues not only in histone proteins but also in many other transcriptional factors including TFIIE and TFIIF, and thus they are also known as factor acetyltransferases [51]. In addition, p300 has been implicated in DNA repair processes because of its involvement in the acetylation of many DNA repair-enzymes such as flap endonuclease 1 (FEN1), DNA polymerase β (Polβ), G/T mismatch-specific DNA glycosylase (TDG) and DNA glycosylases NEIL2 and OGG1 [52–55]. It has been reported that p300 could acetylate both HMGN1 (formerly known as HMG-14) and HMGN2 (formerly known as HMG-17) at multiple sites, whereas PCAF could acetylate only HMGN2 with Lys-2 being the predominant acetylation site [56, 57]. Moreover, Munshi et al. [27] showed that the HAT domains of both CBP and PCAF could acetylate HMGA1a proteins as efficiently as histones in vitro and the preferential acetylation sites were Lys-64 and Lys-70, respectively, according to the in-vitro acetylation of peptide segments of HMGA1 proteins.
To investigate thoroughly the in-vitro acetylation of human HMGA1a and HMGA1b, we first obtained the recombinant proteins, subjected these two proteins to in-vitro acetylation reaction with p300 and PCAF, and monitored the acetylated proteins by MALDI-MS. The MALDI-MS results showed that both p300 and PCAF could induce the acetylation of HMGA1 proteins and multiple lysine residues might be acetylated (data not shown), which made it necessary to carry out further experiments to locate the acetylation sites and to evaluate the levels of acetylation at individual modified lysine residues.
To determine the acetylation sites in HMGA1a protein, we digested the acetylated proteins with trypsin and subjected the digestion mixture to LC-MS/MS analysis on an LTQ linear ion trap mass spectrometer. LC-MS/MS results allowed us to detect both the acetylated (SSQPLASK14QEK) and unacetylated peptides (SSQPLASK14QEK & SSQPLASK14) containing Lys-14 (spectra not shown), which is consistent with our previous in-vivo study [28]. However, we were not able to identify any peptides containing Lys-64 or Lys-70, and these two residues were previously shown to be acetylated by CBP and PCAF, respectively [27].
Since the region harboring Lys-64 and Lys-70 are rich in lysine and arginine residues (Figure 1), the tryptic peptides produced from this region of HMGA1 proteins might be too small to be detected by LC-MS/MS. Therefore, we digested the HMGA1 proteins with Arg-C, which can selectively cleave the amide bonds on the C-terminal side of an arginine residue except for the arginine followed by a proline. Indeed MALDI mass spectrum of the digestion mixture of HMGA1 proteins revealed clearly the presence of both acetylated and unacetylated forms of the peptide GRPKGSKNKGAAKTR (Data not shown). This peptide, however, was still not detectable by ESI-MS/MS analysis, which might be attributed to the formation highly charged ions of this peptide whose m/z values were too low to be chosen for collisional activation.
Although we could detect all the peptides containing Lys-14, Lys-64 and Lys-70, it remained challenging to have a reliable measurement of the level of acetylation at individual lysines for the following reasons. First of all, acetylation of lysine residues in a peptide can alter the ionization efficiency of the peptide. Therefore, the level of acetylation may not be reflected accurately by the relative abundances of ions of the modified and unmodified forms of the peptide. Secondly, because the amide bond on the C-terminal side of an acetylated lysine is no longer susceptible to trypsin cleavage, the acetylation level for Lys-14 has to be calculated based on the ratio of the relative abundance of acetylated peptides (SSQPLASK14QEK) over the sum of the abundances of the two peptides bearing unacetylated Lys-14 (i.e., SSQPLASK14QEK & SSQPLASK14). For such calculation, other than making an assumption that acetylation of Lys-14 does not change the ionization efficiency of a peptide, we also need to assume that the two peptides with different length and amino acid composition share the same ionization efficiency. The deviation from the latter assumption will again lead to inaccurate quantification. Thirdly, although GRPK61GSK64NK66GAAK70TR could be found in both the modified and unmodified forms in the MALDI mass spectrum, the MS/MS of this peptide did not allow us to locate the acetylation sites owing to the poor quality of the spectrum (spectrum not shown).
The above issues, however, can be resolved by applying the quantification method introduced by Smith et al. [42]. As shown in Figure 2, we first treated the acetylated HMGA1 proteins with deuterated acetic anhydride and deuterated acetic acid, which lead to the labeling of all unacetylated lysine residues with a deuterated acetyl moiety. The mass difference between a protiated acetyl group (H3-Ac, 42 Da) and a deuterated acetyl group (D3-Ac, 45 Da) is 3 Da, making it possible to distinguish between the enzyme- and derivatization-induced acetylation. With this chemical derivatization, tryptic peptides containing an originally unacetylated (now labeled with an Ac-D3) or acetylated lysine will have the same length, owing to the inability of trypsin to cleave the amide bond on the C-terminal side of an acetylated lysine, and exhibit comparable ionization efficiency, owing to the similar level of change in ionization efficiency introduced by the addition of an Ac-D3 or an Ac-H3 to the lysine residue. Another advantage of this approach lies in that derivatization leads to a decrease in the charge state of ESI-produced ions of peptides bearing multiple lysine residues. With the above analysis in mind, the acetylation level for each lysine residue can then be calculated from the ion abundance ratio of the protiated and deuterated ions found in MS and/or MS/MS.
Figure 2.
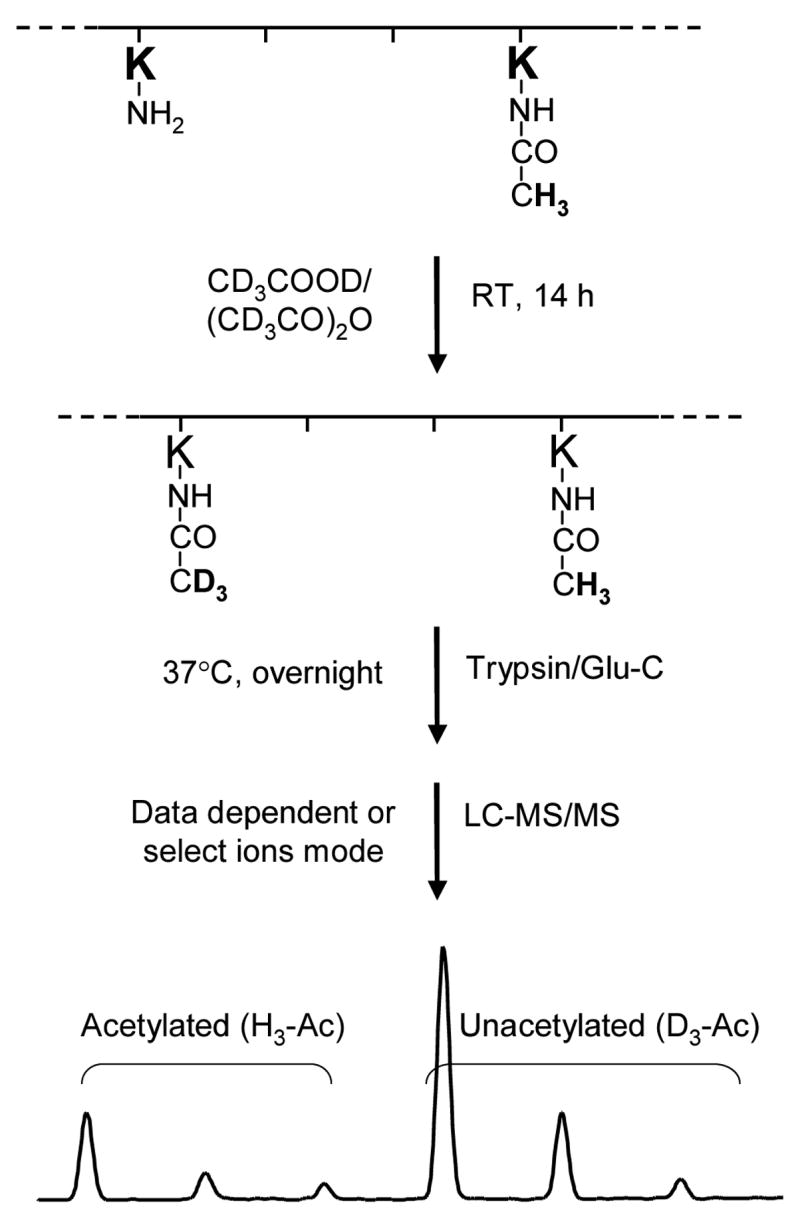
An outline of the experimental approach employed to quantify the in-vitro and in-vivo acetylation of HMGA1a and HMGA1b. HMGA1 proteins were treated with deuterated acetic anhydride and deuterated acetic acid for 14 h. As a result, each of the unacetylated lysine residues was conjugated with a deuterated acetyl group. Digesting the proteins with trypsin or Glu-C produced the desired peptides, in which the acetylated lysine residue (either endogenously acetylated in PC-3 cells or in-vitro acetylated by p300 or PCAF) was tagged with a protiated acetyl moiety (H3-Ac) and the unacetylated lysine residue was attached with a deuterated acetyl group (D3-Ac). Since the mass difference between H3-Ac and D3-Ac is 3 Da, the acetylation level can be determined from the ratio of the abundance of ions bearing H3-Ac over the sum of those bearing H3-Ac and D3-Ac.
Following this approach, we identified five acetylated lysine residues (highlighted in bold italic in Figure 1) residing in 3 peptides (underlined in Figure 1), i.e., SSSKSSQPLASK14QE, GRPKGSK64NK66GAAK70TR, and K73TTTTPGR, of the recombinant HMGA1a that was acetylated in vitro by either p300 or PCAF. The corresponding lysine residues in HMGA1b were also acetylated by PCAF or p300. In this respect, K73TTTTPGR and GRPKGSK64NK66GAAK70TR were identified from the tryptic digestion mixture, whereas SSSKSSQPLASK14QE was identified from the Glu-C digestion mixture.
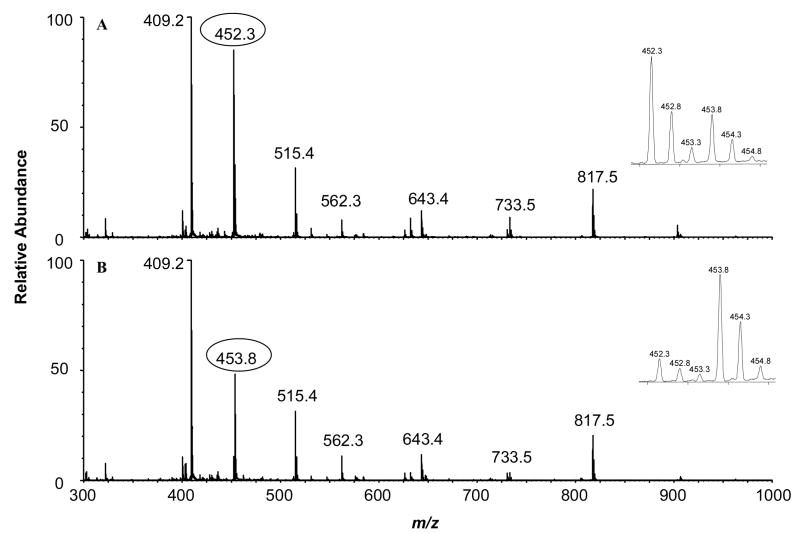
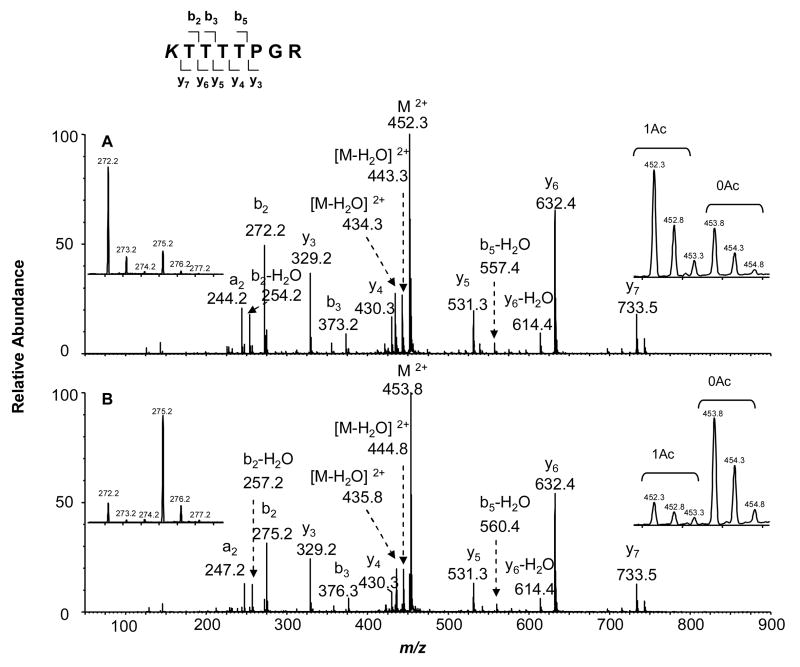
Here we began our discussion with the acetylation of Lys-73. Figures 3 and Figure 4 depict the MS and MS/MS, respectively, of the tryptic peptide K73TTTPGR from HMGA1a that was acetylated in vitro by PCAF or p300. The observed m/z values for the [M+2H]2+ ions of the protiated (m/z 452.3) and deuterated (m/z 453.8) acetyl peptide is consistent with the calculated m/z values. The relative abundances of the ions of m/z 452.3 and m/z 453.8 underscored that much higher level of acetylation was induced by PCAF than by p300. Since there is only one lysine residue in this peptide, the site of acetylation can only be Lys-73, which was also confirmed by MS/MS (Figure 4). The acetylation level for Lys-73 can be quantified from either the relative abundances of the acetylated (m/z 452.3) over unacetylated parent ion (m/z 453.8), or the corresponding ratios for the b-series ions found in MS/MS (e.g., the b2 ion, as shown in the insets on the left of Figure 4A and Figure 4B). Results from triplicate measurements revealed that the average percentages of acetylation induced by PCAF and p300 were 68% and 11%, respectively, indicating that Lys-73 in HMGA1a was more efficiently acetylated by PCAF. In this context, it is worth noting that, under our experimental conditions, we did not observe apparent difference in the retention time for a peptide carrying an H3-Ac and the corresponding peptide bearing a D3-Ac functionality. In addition, we averaged all scans collected in the whole elution time range for the peptide under study to assess quantitatively the levels of acetylation of lysine residues in this and other peptides.
Figure 3.
ESI mass spectra of tryptic peptide K73TTTTPGR from HMGA1a acetylated by histone acetyltransferases PCAF (A) and p300 (B).
Figure 4.
Product-ion spectra of the ESI-produced [M + 2H]2+ ions of tryptic peptide K73TTTTPGR of HMGA1a that was acetylated by PCAF (A) or p300 (B) in vitro. The two insets on the right, showing two groups of peaks, are the expanded mass spectra of the parent ion m/z 452.3 (453.8), corresponding to the unacetylated and monoacetylated forms of this peptide induced by PCAF (top) and p300 (bottom), respectively. The symbol “Ac” designates ions bearing acetyl lysine residues induced by histone acetyltransferases. The two insets on the left are the expanded mass spectra of the b2 ion, corresponding to the acetylated (H3-Ac) and unacetylated (D3-Ac) forms of this product ion, in which the acetylation was induced by PCAF (top) or p300 (bottom). A scheme summarizing the observed fragment ions of this peptide is shown above the mass spectra.
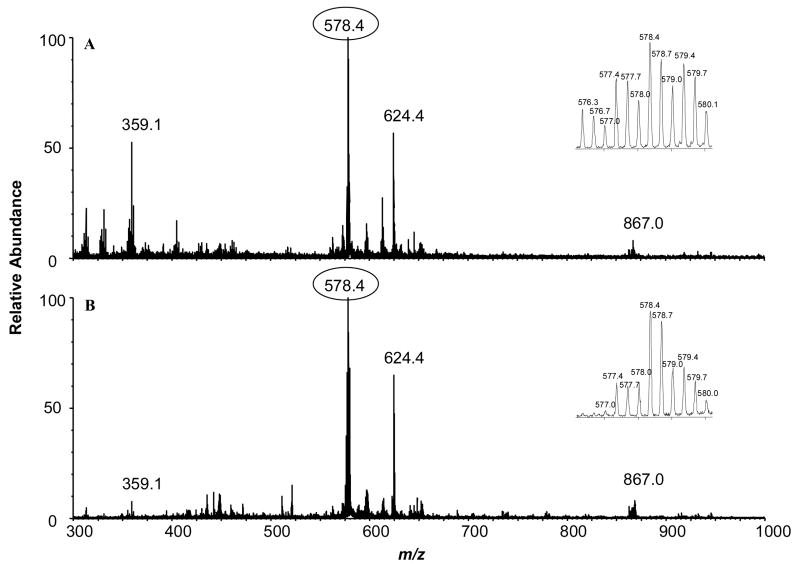
To determine the levels of acetylation of Lys-64, Lys-66, and Lys-70 in GRPKGSK64NK66GAAK70TR, we subjected the tryptic digestion mixtures of p300- and PCAF-acetylated HMGA1a to LC-MS and MS/MS analysis. The MS results for the digestion mixture of HMGA1a acetylated by either p300 or PCAF revealed a set of peaks corresponding to the triply charged ions of the unacetylated, mono-, di- and tri-acetylated peptides (Figure 5). Although the MS data provided useful information about the extent of acetylation of this peptide, we have to rely on MS/MS to determine the acetylation sites and the levels of acetylation of individual lysine residues in this peptide. In this respect, it is worth noting that the window for the isolation of precursor ions was chosen to be wide enough to accommodate all protiated and deuterated forms of the acetylated peptide.
Figure 5.
ESI mass spectra of tryptic peptide GRPKGSK64NK66GAAK70TR from HMGA1a acetylated by histone acetyltransferases PCAF (A) and p300 (B).
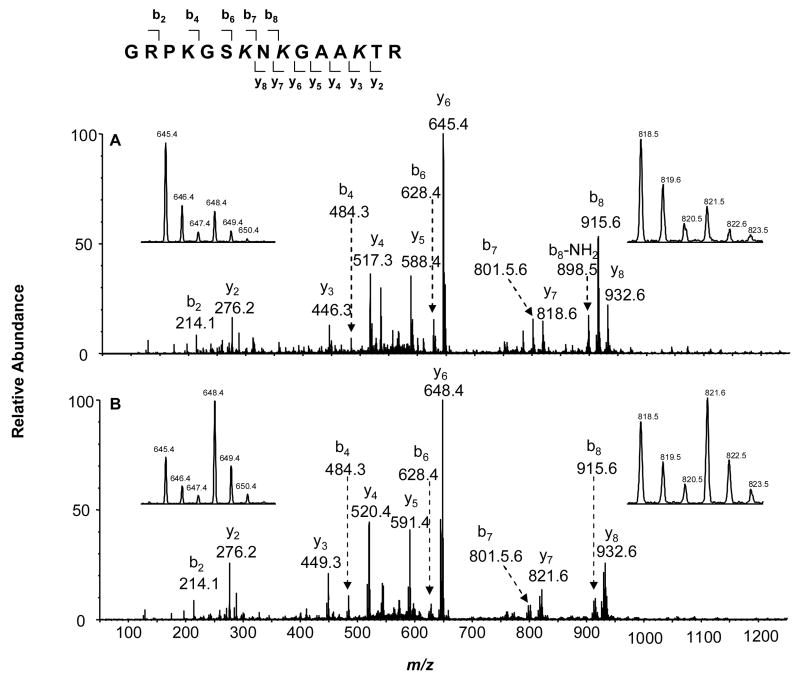
In the product-ion spectra shown in Figure 6, both the protiated and deuterated acetyl moieties were observed to be present for y3, y4 and y6 ions but not for y2, supporting that Lys-70 was partially acetylated by p300 or PCAF and the acetylation level could be determined by the relative abundances of the protiated and deuterated forms of the y3, y4 or y6 ion. Since the y6 ion had higher ion current than the y3 or y4 ion, the y6 ion was used to calculate the acetylation level of Lys-70. We also found that Lys-70 was acetylated more readily by PCAF than by p300 (Figure 6, expanded spectra for y6 ion). Moreover, the y7 and y8 ions exhibited higher abundance ratios of protiated ion (acetylated) over deuterated ion (unacetylated) than that of y6, suggesting that Lys-66 was also acetylated by p300 and PCAF. The acetylation level was determined based on the y7 and y6 ions using the equation described previously [42]. MS/MS results also support that Lys-61 in this peptide was not acetylated because the b4 ion only carries the D3-Ac-, but not the H3-Ac-labeled lysine. On the other hand, we found that the b7 and b8 ions encompass both the D3-Ac- and the H3-Ac-labeled lysine, supporting that Lys-64 is acetylated. The percentage of acetylation for this residue was determined from the b7 ion.
Figure 6.
Product-ion spectra of the ESI-produced [M + 2H]2+ ion of the tryptic peptide GRPKGSK64NK66GAAK70TR from HMGA1a that was acetylated in vitro by PCAF (A) and p300 (B). The four insets are the expanded mass spectra of y6 (left) and y7 (right) ions, in which the acetylation was induced by PCAF (top) and p300 (bottom). A scheme summarizing the observed fragment ions of this peptide is shown above the mass spectra.
The peptides containing Lys-14 was not detected from LC-MS/MS analysis of the tryptic digestion mixture of the HMGA1a that has been acetylated by p300 or PCAF and derivatized with deuterated acetic acid and deuterated acetic anhydride. On the grounds that only the amide bonds on the C-terminal side of arginine residues can be cleaved after all the lysine residues are acetylated enzymatically or chemically, Lys-14 is expected to be present in the tryptic peptide harboring residues 1–23, which might be somewhat too long for LC-MS/MS identification. After inspecting the sequence of HMGA1 proteins, we chose to employ Glu-C for the digestion, which gave rise to the peptide SSSKSSQPLASKQE. MS/MS of the [M+2H]2+ ion of this peptide facilitates us to conclude that Lys-14 in this peptide was acetylated and the acetylation level of this residue can be calculated either based on the [M+2H]2+ ion found in the MS or appropriate fragment ions (the ion y7 was chosen here due to its high abundance) observed in MS/MS (Data not shown).
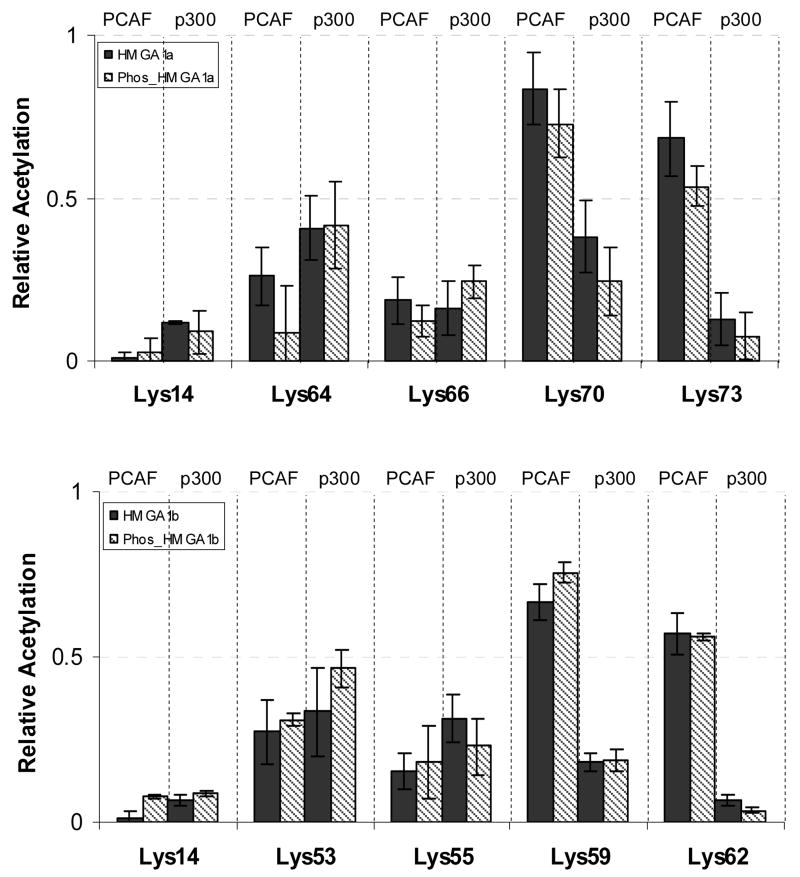
The quantitative results from triplicate measurements for the acetylation levels of all five identified lysine residues in HMGA1a and HMGA1b are summarized in Figure 7. It turned out that p300 and PCAF exhibited different substrate specificity profiles. In particular, the preference of p300 acetylation followed the order of Lys-64~Lys-70 > Lys-66 > Lys-14~Lys73, whereas the selectivity of PCAF acetylation followed the sequence of Lys-70~Lys-73 > Lys-64~Lys-66 > Lys-14. On the other hand, the acetylation profiles of HMGA1a and HMGA1b, induced by either PCAF or p300, showed remarkable similarity, underlying that acetylation might not play a role in the different functional roles assumed by HMGA1a and HMGA1b.
Figure 7.
Histograms showing the fraction of in-vitro acetylation at Lys-14, Lys-64, Lys-66, Lys-70 and Lys-73 of recombinant HMGA1a and phosphorylated HMGA1a (phos-HMGA1a) catalyzed by both p300 and PCAF (top panel), and the fraction of in-vitro acetylation at Lys-14, Lys-53, Lys-55 Lys-59 and Lys-62 of recombinant HMGA1b and phosphorylated HMGA1b (phos-HMGA1b) catalyzed by both p300 and PCAF (bottom panel).
The effect of C-terminal phosphorylation on the acetylation of HMGA1 proteins
Protein kinase CK2 is known to phosphorylate the acidic C-terminal tail of HMGA1a proteins at Ser-102 and Ser-103 both in vitro and in vivo [15, 58], linking intracellular signaling to phosphorylation events occurring in the nucleus [16]. Prior research demonstrated that HMGA1 proteins were both phosphorylated and acetylated in vivo [28, 43], and it has been suggested that HMGA1a proteins, like histones, may exhibit a biochemical modification “code” which is associated with the cellular functions of these two proteins [7, 9, 19]. Therefore, it is important to examine whether C-terminal constitutive phosphorylation of HMGA1 induced by protein kinase CK2 may affect its acetylation. To this end, we carried out the in-vitro phosphorylation of HMGA1 proteins with protein kinase CK2 [45] followed by in-vitro acetylation with histone acetyltransferases p300 and PCAF. We then assessed the acetylation of the two proteins by using the same strategy as discussed above, and it turned out that the C-terminal phosphorylation did not affect appreciably the acetylation of the HMGA1 proteins (Figure 7).
Acetylation of HMGA1 proteins isolated from PC-3 cells
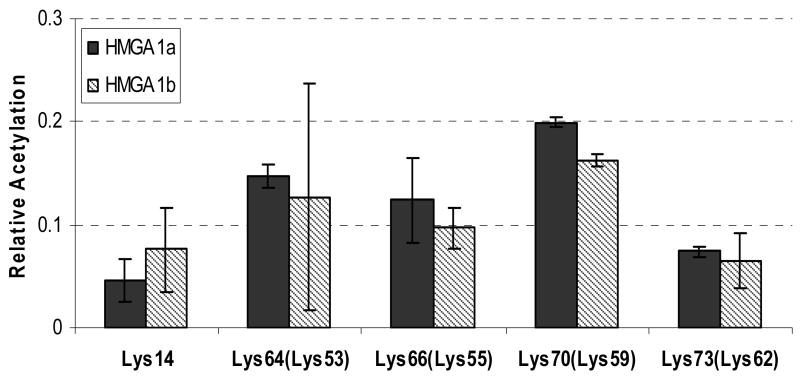
After identifying the sites and quantifying the levels of acetylation in these two proteins induced by p300 and PCAF in vitro, we set out to investigate whether these lysine residues are also acetylated in vivo. To this end, we purified the HMGA1a and HMGA1b proteins from PC-3 human prostate cancer cells and assessed the acetylation of these two proteins by using the same experimental procedures as discussed above. Our results illustrated that all five residues that are susceptible to acetylation in vitro were also detected to be acetylated in vivo (Figure 8). In this respect, less than 10% of Lys-14 and Lys-73 was acetylated in vivo, whereas the average percentages of acetylation for Lys-70, Lys-64 and Lys-66 in HMGA1a were 20%, 15% and 12%, respectively. In this regard, similar distribution in acetylation levels was found for HMGA1b (Figure 8). The large error bar for the extent of acetylation of Lys-51 in HMGA1b could be due to the low intensity for the ion used for quantifying the acetylation level of this residue.
Figure 8.
Fractions of acetylation at residues Lys-14, Lys-64, Lys-66, Lys-70, Lys-73 of HMGA1a and at residues Lys-14, Lys-53, Lys-55, Lys-59, Lys-62 of HMGA1b purified from PC-3 human prostate cancer cells.
DISCUSSION
The specific and controlled acetylation of lysine residues in histones plays an important role in the modulation of chromatin structure associated with transcriptional activation [59, 60]. HMGA1 proteins, like histone proteins, are susceptible to acetylation both in vivo and in vitro [8, 26–28], and acetylation represents one of the prominent PTMs of HMGA1 proteins in living cells [1].
In the present study, we examined systematically both the sites and the degrees of acetylation of HMGA1 proteins in vitro and in PC-3 cells by using a mass spectrometry-based approach. In particular, we employed a chemical derivatization method, first introduced by Smith and coworkers [42], for the quantification of the levels of acetylation. There are three advantages of this quantification strategy over other methods for quantifying HMGA1a acetylation. First of all, peptides bearing an unmodified lysine residues, after being labeled with deuterated acetyl moiety (D3-Ac), or an acetylated lysine residues (H3-Ac) have very similar, if not identical, ionization efficiencies. Secondly, chemical acetylation decreased the charge state of electrospray-produced ions of the digested peptides containing multiple lysine residues, which made it possible to detect GRPKGSK64NK66GAAK70TR by LC-ESI-MS and MS/MS. Moreover, the D3-Ac-modified lysine introduced by derivatization blocked the cleavage of the amide bond on the C-terminal side of the modified lysine residues in the same way as the H3-Ac-modified lysine induced by p300 or PCAF. Therefore, tryptic peptides bearing the originally unmodified and acetylated lysine residues share the same sequence, which allows for the quantification of the acetylation level. Thirdly, this approach allowed for the quantification of the levels of individual lysine residues in a peptide carrying multiple acetylated lysines.
Several conclusions can be drawn from our quantification measurements. First, HMGA1a and HMGA1b exhibited a strikingly similar pattern of acetylation induced by either p300 or PCAF. In this regard, it was suggested that HMGA1a and HMGA1b, despite their structural similarity, may have distinct biological functions [8, 10] and, as a result, may undergo different posttranslational modifications [9, 20, 24]. To explore any possible differences between HMGA1a and HMGA1b in acetylation, we investigated thoroughly the p300- and PCAF-catalyzed in-vitro acetylation of the recombinant proteins and the recombinant proteins phosphorylated by protein kinase CK2. All the results suggested that both the sites and the levels of acetylation induced by p300 or PCAF were remarkably similar for HMGA1a and HMGA1b. Secondly, p300 and PCAF acetylated the HMGA1 proteins in vitro with different profiles of site specificity. Previous in-vitro acetylation assay [27] suggested that, except for the major acetylation sites of Lys-64 induced by CBP and Lys-70 induced by PCAF, many other lysine residues can also be acetylated, especially by PCAF. We, however, detected only five acetylation sites, which might be attributed to the fact that protein substrates, rather than peptide substrates, were employed to examine the site preferences of histone acetyltransferases in the present study. Thirdly, G/SK may not always be the consensus acetylation motif induced by p300 as previously reported [61]. Our in-vitro acetylation data revealed that, among the five lysine residues that were acetylated, only Lys-14 and Lys-64 reside in this motif. Moreover, Lys-6 is located in this G/SK motif in HMGA1a and HMGA1b, however, it was not acetylated by p300.
Our data also demonstrated that C-terminal phosphorylation of HMGA1 proteins did not exert obvious effect on the acetylation of these two proteins induced by p300 or PCAF. We reason that the acidic C-terminal tail of HMGA1 proteins is somewhat remote from the lysine residues that are to be acetylated and, therefore, the incorporation of additional negative charge to the C-terminal motif does not affect the access of p300 or PCAF to the lysine residues to be acetylated.
We further demonstrated that all five lysine residues in HMGA1 proteins that could be acetylated by p300 and PCAF in-vitro were also acetylated in PC-3 human prostate cancer cells. Reversible acetylation of Lys-64 and Lys-70 modulates the transcription of IFN-β gene by regulating the destabilization and formation, respectively, of an enhancesome on the promoter region of the IFN-β gene [26, 27]. Lys-64 and Lys-70 in HMGA1a were found to be acetylated in nonmetastatic and moderately metastatic breast cancer cells such as MCF-7/Tet-off cells and HA7C cells, but not in highly metastatic HA8A breast cancer cells [19]. In addition, Lys-14 in HMGA1 proteins was observed to be acetylated in PC-3 [28] and HA7C cells [19], whereas the same residue was monomethylated in HMGA1a in non-metastatic MCF-7/Tet-off cells. The biological implications of the acetylation of these lysine residues remain to be established.
Our quantification data revealed that HMGA1a and HMGA1b shared a similar acetylation pattern in living cells, indicating that acetylation was not responsible for the PTM-associated different functions of HMGA1a and HMGA1b. In-vitro acetylation results showed that Lys-14 and Lys-64 were preferentially acetylated by p300, whereas Lys-70 and Lys-73 were favorable acetylation sites induced by PCAF. On the grounds that both Lys-64 and Lys-70 exhibited relatively high and comparable levels of acetylation, we suspect that both p300 and PCAF might be involved in the in-vivo acetylation of HMGA1 proteins in PC-3 cells, which was consistent with the fact that both CBP and PCAF co-immunoprecipitate with HMGA1 proteins [27]. However, we cannot exclude the possibility that other acetyltransferases might also be involved in the acetylation of HMGA1 proteins.
Acknowledgments
The authors want to thank the National Institutes of Health for supporting this research (grant No. CA101864).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 2.Reeves R, Nissen MS. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 3.Elton TS, Nissen MS, Reeves R. Specific A. T DNA sequence binding of RP-HPLC purified HMG-I. Biochem Biophys Res Commun. 1987;143:260–265. doi: 10.1016/0006-291x(87)90659-0. [DOI] [PubMed] [Google Scholar]
- 4.Johnson KR, Lehn DA, Reeves R. Alternative processing of mRNAs encoding mammalian chromosomal high-mobility-group proteins HMG-I and HMG-Y. Mol Cell Biol. 1989;9:2114–2123. doi: 10.1128/mcb.9.5.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson KR, Lehn DA, Elton TS, Barr PJ, Reeves R. Complete murine cDNA sequence, genomic structure, and tissue expression of the high mobility group protein HMG-I(Y) J Biol Chem. 1988;263:18338–18342. [PubMed] [Google Scholar]
- 6.Reeves R, Beckerbauer L. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim Biophys Acta. 2001;1519:13–29. doi: 10.1016/s0167-4781(01)00215-9. [DOI] [PubMed] [Google Scholar]
- 7.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 8.Banks GC, Li Y, Reeves R. Differential in vivo modifications of the HMGI(Y) nonhistone chromatin proteins modulate nucleosome and DNA interactions. Biochemistry. 2000;39:8333–8346. doi: 10.1021/bi000378+. [DOI] [PubMed] [Google Scholar]
- 9.Edberg DD, Adkins JN, Springer DL, Reeves R. Dynamic and differential in vivo modifications of the isoform HMGA1a and HMGA1b chromatin proteins. J Biol Chem. 2005;280:8961–8973. doi: 10.1074/jbc.M407348200. [DOI] [PubMed] [Google Scholar]
- 10.Reeves R, Edberg DD, Li Y. Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol Cell Biol. 2001;21:575–594. doi: 10.1128/MCB.21.2.575-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lund T, Holtlund J, Laland SG. On the phosphorylation of low molecular mass HMG (high mobility group) proteins in Ehrlich ascites cells. FEBS Lett. 1985;180:275–279. doi: 10.1016/0014-5793(85)81085-1. [DOI] [PubMed] [Google Scholar]
- 12.Reeves R, Langan TA, Nissen MS. Phosphorylation of the DNA-binding domain of nonhistone high-mobility group I protein by cdc2 kinase: reduction of binding affinity. Proc Natl Acad Sci USA. 1991;88:1671–1675. doi: 10.1073/pnas.88.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lund T, Laland SG. The metaphase specific phosphorylation of HMG I. Biochem Biophys Res Commun. 1990;171:342–347. doi: 10.1016/0006-291x(90)91399-d. [DOI] [PubMed] [Google Scholar]
- 14.Xiao DM, Pak JH, Wang X, Sato T, Huang FL, Chen HC, Huang KP. Phosphorylation of HMG-I by protein kinase C attenuates its binding affinity to the promoter regions of protein kinase C gamma and neurogranin/RC3 genes. J Neurochem. 2000;74:392–399. doi: 10.1046/j.1471-4159.2000.0740392.x. [DOI] [PubMed] [Google Scholar]
- 15.Palvimo J, Linnala-Kankkunen A. Identification of sites on chromosomal protein HMG-I phosphorylated by casein kinase II. FEBS Lett. 1989;257:101–104. doi: 10.1016/0014-5793(89)81796-x. [DOI] [PubMed] [Google Scholar]
- 16.Wang DZ, Ray P, Boothby M. Interleukin 4-inducible phosphorylation of HMG-I(Y) is inhibited by rapamycin. J Biol Chem. 1995;270:22924–22932. doi: 10.1074/jbc.270.39.22924. [DOI] [PubMed] [Google Scholar]
- 17.Schwanbeck R, Gymnopoulos M, Petry I, Piekielko A, Szewczuk Z, Heyduk T, Zechel K, Wisniewski JR. Consecutive steps of phosphorylation affect conformation and DNA binding of the chironomus high mobility group A protein. J Biol Chem. 2001;276:26012–26021. doi: 10.1074/jbc.M011053200. [DOI] [PubMed] [Google Scholar]
- 18.Piekielko A, Drung A, Rogalla P, Schwanbeck R, Heyduk T, Gerharz M, Bullerdiek J, Wisniewski JR. Distinct organization of DNA complexes of various HMGI/Y family proteins and their modulation upon mitotic phosphorylation. J Biol Chem. 2001;276:1984–1992. doi: 10.1074/jbc.M004065200. [DOI] [PubMed] [Google Scholar]
- 19.Edberg DD, Bruce JE, Siems WF, Reeves R. In vivo posttranslational modifications of the high mobility group A1a proteins in breast cancer cells of differing metastatic potential. Biochemistry. 2004;43:11500–11515. doi: 10.1021/bi049833i. [DOI] [PubMed] [Google Scholar]
- 20.Sgarra R, Diana F, Bellarosa C, Dekleva V, Rustighi A, Toller M, Manfioletti G, Giancotti V. During apoptosis of tumor cells HMGA1a protein undergoes methylation: identification of the modification site by mass spectrometry. Biochemistry. 2003;42:3575–3585. doi: 10.1021/bi027338l. [DOI] [PubMed] [Google Scholar]
- 21.Sgarra R, Diana F, Rustighi A, Manfioletti G, Giancotti V. Increase of HMGA1a protein methylation is a distinctive characteristic of leukaemic cells induced to undergo apoptosis. Cell Death Differ. 2003;10:386–389. doi: 10.1038/sj.cdd.4401184. [DOI] [PubMed] [Google Scholar]
- 22.Miranda TB, Webb KJ, Edberg DD, Reeves R, Clarke S. Protein arginine methyltransferase 6 specifically methylates the nonhistone chromatin protein HMGA1a. Biochem Biophys Res Commun. 2005;336:831–835. doi: 10.1016/j.bbrc.2005.08.179. [DOI] [PubMed] [Google Scholar]
- 23.Sgarra R, Lee J, Tessari MA, Altamura S, Spolaore B, Giancotti V, Bedford MT, Manfioletti G. The AT-hook of the chromatin architectural transcription factor high mobility group A1a is arginine-methylated by protein arginine methyltransferase 6. J Biol Chem. 2006;281:3764–3772. doi: 10.1074/jbc.M510231200. [DOI] [PubMed] [Google Scholar]
- 24.Zou Y, Wang Y. Tandem mass spectrometry for the examination of the posttranslational modifications of high-mobility group A1 proteins: symmetric and asymmetric dimethylation of Arg25 in HMGA1a protein. Biochemistry. 2005;44:6293–6301. doi: 10.1021/bi0475525. [DOI] [PubMed] [Google Scholar]
- 25.Zou Y, Wang Y. Mass spectrometric analysis of high-mobility group proteins and their post-translational modifications in normal and cancerous human breast tissues. J Proteome Res. 2007;6 doi: 10.1021/pr070072q. in press. [DOI] [PubMed] [Google Scholar]
- 26.Munshi N, Agalioti T, Lomvardas S, Merika M, Chen G, Thanos D. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science. 2001;293:1133–1136. doi: 10.1126/science.293.5532.1133. [DOI] [PubMed] [Google Scholar]
- 27.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 28.Jiang X, Wang Y. Acetylation and phosphorylation of high-mobility group A1 proteins in PC-3 human tumor cells. Biochemistry. 2006;45:7194–7201. doi: 10.1021/bi060504v. [DOI] [PubMed] [Google Scholar]
- 29.Meng F, Forbes AJ, Miller LM, Kelleher NL. Detection and localization of protein modifications by high resolution tandem mass spectrometry. Mass Spectrom Rev. 2005;24:126–134. doi: 10.1002/mas.20009. [DOI] [PubMed] [Google Scholar]
- 30.Larsen MR, Trelle MB, Thingholm TE, Jensen ON. Analysis of posttranslational modifications of proteins by tandem mass spectrometry. Biotechniques. 2006;40:790–798. doi: 10.2144/000112201. [DOI] [PubMed] [Google Scholar]
- 31.Goshe MB, Smith RD. Stable isotope-coded proteomic mass spectrometry. Curr Opin Biotechnol. 2003;14:101–109. doi: 10.1016/s0958-1669(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 32.Julka S, Regnier F. Quantification in proteomics through stable isotope coding: a review. J Proteome Res. 2004;3:350–363. doi: 10.1021/pr0340734. [DOI] [PubMed] [Google Scholar]
- 33.Tao WA, Aebersold R. Advances in quantitative proteomics via stable isotope tagging and mass spectrometry. Curr Opin Biotechnol. 2003;14:110–118. doi: 10.1016/s0958-1669(02)00018-6. [DOI] [PubMed] [Google Scholar]
- 34.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci USA. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 36.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 37.Lemmel C, Weik S, Eberle U, Dengjel J, Kratt T, Becker HD, Rammensee HG, Stevanovic S. Differential quantitative analysis of MHC ligands by mass spectrometry using stable isotope labeling. Nat Biotechnol. 2004;22:450–454. doi: 10.1038/nbt947. [DOI] [PubMed] [Google Scholar]
- 38.Goshe MB, Veenstra TD, Panisko EA, Conrads TP, Angell NH, Smith RD. Phosphoprotein isotope-coded affinity tags: application to the enrichment and identification of low-abundance phosphoproteins. Anal Chem. 2002;74:607–616. doi: 10.1021/ac015528g. [DOI] [PubMed] [Google Scholar]
- 39.Goshe MB, Conrads TP, Panisko EA, Angell NH, Veenstra TD, Smith RD. Phosphoprotein isotope-coded affinity tag approach for isolating and quantitating phosphopeptides in proteome-wide analyses. Anal Chem. 2001;73:2578–2586. doi: 10.1021/ac010081x. [DOI] [PubMed] [Google Scholar]
- 40.Xiong L, Andrews D, Regnier F. Comparative proteomics of glycoproteins based on lectin selection and isotope coding. J Proteome Res. 2003;2:618–625. doi: 10.1021/pr0340274. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 42.Smith CM, Gafken PR, Zhang Z, Gottschling DE, Smith JB, Smith DL. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Anal Biochem. 2003;316:23–33. doi: 10.1016/s0003-2697(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 43.Elton TS, Reeves R. Purification and postsynthetic modifications of Friend erythroleukemic cell high mobility group protein HMG-I. Anal Biochem. 1986;157:53–62. doi: 10.1016/0003-2697(86)90195-8. [DOI] [PubMed] [Google Scholar]
- 44.Reeves R. HMGA proteins: isolation, biochemical modifications, and nucleosome interactions. Methods Enzymol. 2004;375:297–322. doi: 10.1016/s0076-6879(03)75020-4. [DOI] [PubMed] [Google Scholar]
- 45.Zou Y, Webb K, Perna AD, Zhang Q, Clarke S, Wang Y. A mass spectrometric study on the in vitro methylation of HMGA1a and HMGA1b proteins by PRMTs: methylation specificity, the effect of binding to AT-rich duplex DNA and the effect of C-terminal phosphorylation. Biochemistry. 2007;46 doi: 10.1021/bi6024897. in press. [DOI] [PubMed] [Google Scholar]
- 46.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 47.Brownell JE, Allis CD. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 49.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 50.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 51.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhakat KK, Hazra TK, Mitra S. Acetylation of the human DNA glycosylase NEIL2 and inhibition of its activity. Nucleic Acids Res. 2004;32:3033–3039. doi: 10.1093/nar/gkh632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhakat KK, Izumi T, Yang SH, Hazra TK, Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 2003;22:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hasan S, Stucki M, Hassa PO, Imhof R, Gehrig P, Hunziker P, Hubscher U, Hottiger MO. Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol Cell. 2001;7:1221–1231. doi: 10.1016/s1097-2765(01)00272-6. [DOI] [PubMed] [Google Scholar]
- 55.Bhakat KK, Mokkapati SK, Boldogh I, Hazra TK, Mitra S. Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol Cell Biol. 2006;26:1654–1665. doi: 10.1128/MCB.26.5.1654-1665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herrera JE, Sakaguchi K, Bergel M, Trieschmann L, Nakatani Y, Bustin M. Specific acetylation of chromosomal protein HMG-17 by PCAF alters its interaction with nucleosomes. Mol Cell Biol. 1999;19:3466–3473. doi: 10.1128/mcb.19.5.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bergel M, Herrera JE, Thatcher BJ, Prymakowska-Bosak M, Vassilev A, Nakatani Y, Martin B, Bustin M. Acetylation of novel sites in the nucleosomal binding domain of chromosomal protein HMG-14 by p300 alters its interaction with nucleosomes. J Biol Chem. 2000;275:11514–11520. doi: 10.1074/jbc.275.15.11514. [DOI] [PubMed] [Google Scholar]
- 58.Ferranti P, Malorni A, Marino G, Pucci P, Goodwin GH, Manfioletti G, Giancotti V. Mass spectrometric analysis of the HMGY protein from Lewis lung carcinoma. Identification of phosphorylation sites. J Biol Chem. 1992;267:22486–22489. [PubMed] [Google Scholar]
- 59.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 60.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 61.Bannister AJ, Miska EA, Gorlich D, Kouzarides T. Acetylation of importin-alpha nuclear import factors by CBP/p300. Curr Biol. 2000;10:467–470. doi: 10.1016/s0960-9822(00)00445-0. [DOI] [PubMed] [Google Scholar]