Abstract
Background/aims: Current in vitro drug sensitivity tests have limitations and disadvantages. This study investigated the use of gene expression data to predict the sensitivity of pancreatic cancers to gemcitabine. Materials and methods: Cancer cells isolated from 14 pancreatic cancer patients were tested in vitro for gemcitabine sensitivity using the collagen droplet drug sensitivity test (CD-DST). On the basis of this test, 9 of the 14 cancers were identified as either gemcitabine-sensitive or gemcitabine-resistant. Total RNA was extracted from each of those nine cancers and used as a template to synthesize Cy3-labeled cDNA. Pancreatic RNA extracted from six normal individuals was used as a control. Labeled probes were hybridized to an Atlas Glass Human 1.0 Microarray chip, after which the chips were washed and scanned, and the data were analyzed using Microsoft Excel-embedded software. The expression profiles of selected genes were confirmed using real-time PCR analysis. Results: Statistical analysis of the microarray data showed that four genes were differentially expressed in gemcitabine-sensitive cancers: microsomal glutathione S-transferase 1 (GSTT1), topoisomerase II alpha (TOP2A), caspase 3, and ATP-binding cassette and subfamily C member 2 (ABCC2). More than 20 other genes were additionally identified as possible candidate genes associated with drug resistance. Conclusions: Expression of drug resistance-related genes appeared to predict whether a cancer was gemcitabine-sensitive or -resistant. Further study will enable a drug resistance scoring system to be established on the basis of gene expression. Such a system will allow more efficient application of chemotherapy.
Keywords: Gemcitabine, pancreatic cancer, microarray, drug resistance
Introduction
Treatment of carcinoma of the exocrine pancreas is a major problem, with approximately 80% of patients presenting with unresectable disease due to metastases and/or local invasion 1. Despite many advances in solid tumor therapies over recent decades, unresectable pancreatic cancer continues to have a median survival time of only 3–6 months. The development of gemcitabine, a deoxycytidine analog related to cytarabine, has prompted renewed interest in developing cytotoxic therapies for pancreatic cancer. Although gemcitabine is a well-tolerated drug and ideal for palliation of symptomatic cancer, the efficacy rates remain at only 20–30%. However, in some cases tumors respond well to this treatment and patients experience long survival times.
Since the characteristics of pancreatic cancer can vary between individuals, chemotherapy should ideally be tailored to each patient based on the nature of their particular disease. The detection of potentially chemo-sensitive tumors would significantly improve response rates and facilitate the selection of effective individualized regimens. Developing a method of assessing the likely effectiveness of anticancer drugs using resected or biopsied materials before treatment is likely to avoid unnecessary treatment.
A number of tests to determine the chemo-sensitivity of cancers to particular drugs have been developed, including the nude mouse method, subrenal capsule assay (SRC), human tumor clonogenic assay (HTCA), thymidine incorporation assay (TIA), succinic dehydrogenase inhibition test (SDI test), and the MTT assay 2. However, none of these methods has been widely adopted in clinical practice for various reasons, including low success rates for primary culture, a large number of cells being required for testing, difficulty in ruling out the effect of contaminating fibroblasts, assays taking more than a week, and skilled technicians being required. Although Kobayashi et al. developed a collagen-gel droplet embedded culture drug sensitivity test (CD-DST) which avoids some of these shortcomings 3, the method still requires significant quantities of fresh tissue, usually more than 0.5 cm3.
Pancreatic cancers contain a high proportion of fibroblasts and connective tissue, making it difficult to obtain sufficient cancer tissue for primary culture testing. The success rate of such cultures remains at about 60–80%. However, the development of microarray technology has made it possible to evaluate pancreatic cancers using less tissue than required for CD-DST, and with a higher success rate. Assersohn et al. showed breast cancer gene expression profiles in 15% of patients using tissue from fine needle aspirates and cDNA microarrays 4. This microarray approach is likely to become more common with increasingly sensitive scanning techniques and validated amplification techniques.
The present study investigated the use of gene expression microarray technology for predicting the chemo-sensitivity of pancreatic cancers.
Materials and methods
The study involved 14 patients with biopsy-proven ductal adenocarcinoma of the pancreas admitted to the Jichi Medical School Hospital (Tochigi, Japan) between January 2001 and December 2003 (Table I). We obtained approval from the ethics committee in Jichi Medical University, and documented informed consents from all patients. A laparotomy was performed to obtain 250–1000 mg of fresh pancreatic cancer tissue.
Table I. Profiles and chemo-sensitivities of the 14 pancreatic cancer patients.
| Case | Age (years)/gender | Location | Operation | T/C ratio | Sensitive/resistant | Outcome | Duration (M) | Chemotherapy (M) |
|---|---|---|---|---|---|---|---|---|
| 1 | 69/F | Head | PpPD | 20.8 | Sensitive | Dead | 15.9 | 7 |
| 2 | 62/M | Head | Unresectable | None | Unknown | Dead | 6.8 | 2 |
| 3 | 75/M | Head | Unresectable | 74.4 | Resistant | Dead | 14.3 | 8 |
| 4 | 61/M | Head | Unresectable | 89.4 | Resistant | Dead | 8.1 | 4 |
| 5 | 57/M | Head | Unresectable | 76.2 | Resistant | Dead | 15.5 | 7 |
| 6 | 59/F | Head | PpPD | 64.6 | Resistant | Dead | 15.6 | 11 |
| 7 | 70/F | Body | DP | None | Unknown | Dead | 11.3 | 2 |
| 8 | 78/F | Head | PpPD | None | Unknown | Alive | 41.2 | 12 |
| 9 | 72/F | Head | PpPD | 76.2 | Resistant | Dead | 10.1 | 5 |
| 10 | 75/F | Tail | DP | None | Unknown | Dead | 12.3 | 7 |
| 11 | 77/F | Head | PpPD | 30 | Sensitive | Alive | 42.0 | 12 |
| 12 | 64/M | Head | Unresectable | 33.3 | Sensitive | Dead | 16.1 | 11 |
| 13 | 65/M | Body | DP | None | Unknown | Dead | 24.4 | 12 |
| 14 | 74/M | Head | Unresectable | 38.7 | Sensitive | Dead | 16.6 | 10 |
PpPD, pylorus-preserving pancreatoduodenectomy; DP, distal pancreatectomy.
CD-DST chemo-sensitivity tests were performed using a human tumor cell primary culture system kit (Primaster, Nitta Gelatin, Osaka, Japan) 5. Briefly, fresh surgical specimens from pancreatic cancers were cut into small pieces aseptically and suspended in Hanks's balanced saline solution (HBSS). Cells were dispersed by incubating tissue at 37°C for 1–3 h in a 0.1% cell dispersion enzyme solution (EZ, Nitta Gelatin). Cells were then centrifuged at 900 g for 3 min and the pellet was resuspended in PCM-1 medium (Nitta Gelatin), and the suspension filtered through an 80 µm pore nylon mesh. After preliminary culture in a collagen gel-coated flask in a CO2 incubator at 37°C for 24–48 h, 3×103 cells were added to a 30 µl collagen gel droplet. Cells were cultured in DF medium (Nissui Pharmaceutical Co., Tokyo, Japan) containing 10% fetal bovine serum (Gibco BRL, Grand Island, NY, USA) with or without 0.4 mg/ml gemcitabine for 24 h. Quantification of the total volume of a cell colony, utilizing differences in the growth morphologies of tumor cells and fibroblasts, was determined using an image analysis method 6. The effect of gemcitabine was determined by calculating the ratio of the total colony volume of cells with (T) and without (C) gemcitabine. Cells with a T/C ratio < 50% were considered gemcitabine-sensitive, while those with a ratio > 50% were considered gemcitabine-resistant.
Gene expression profiles were evaluated using microarray techniques. Briefly, purified total RNA from frozen samples was isolated using Atlas Glass Total RNA Isolation Kits (Clontech, Palo Alto, USA) according to the manufacturer's protocols. cDNA was synthesized using BD Atlas PowerScript Fluorescent Labeling Kits, and the resultant Cy3-labeled (Amersham Pharmacia Biotech, Bucks, UK) double-stranded cDNA was purified using QIAquick PCR Purification Kits (QIAGEN Valencia). Cy3-labeled cDNA synthesized from a pool of normal pancreatic RNA (BioChain Institute, Hay ward) was used as a control. Cy3-labeled cDNA was hybridized to a BD Atlas Glass Human 1.0 Microarray (Clontech) in a water bath at 50°C for 16 h. Chips were then washed in four high-volume wash chambers (Clontech). Using a GMS 418 Array Scanner (Takara, Tokyo) and accompanying software, fluorescence intensities for dyes Cy3 were determined and subtraction of local background values for individual spots was performed. The data were exported to Microsoft Excel spreadsheets for analysis. To normalize for the amount of total RNA on each chip, the sample/control ratio for the expression of each gene was adjusted so that the averaged Cy3:Cy3 ratio of seven housekeeping genes was given the value of 1.0, and the data then underwent log2 transformation. To identify genes that were differentially expressed between drug-sensitive and drug-resistant cancers, the Excel-embedded statistical software ‘Analyse-it’ was used to calculate the U and p values for the Mann–Whitney analysis of each gene. A difference in gene expression was considered significant if the p value was < 0.05.
Differential expression of genes identified by microarray analysis was confirmed using real-time PCR analysis and specific primers (Table II). Total RNA used for the microarray analysis was also used for the real-time PCR analysis. Primers were designed for the genes of interest using GENETYX-WIN software (Software Development Corporation, Tokyo, Japan), and then PCR conditions were optimized for each pair of primers (QuantiTec SYBR Green PCR Kit, Qiagen KK, Tokyo, Japan). First strand cDNA was then synthesized from 2 µg total RNA (Superscript First Strand cDNA Synthesis Kit), and 1 µl RT-PCR product was used in real-time PCR assays under optimized reaction conditions. The 50 µl reaction mixture comprised 25 µl SYBR Green PCR Master Mix, 1 µl sense primer, 1 µl antisense primer, 1 µl cDNA, 0.5 µl uracil-N-glycosylase, and 21.5 µl RNase-free water. The real-time cycler conditions were 50°C for 2 min, 95°C for 10 min, 94°C for 15 s, optimized annealing temperature for 30 s, 72°C for 30 s, 50 cycles. β-Actin expression was used as a control for normalizing the amounts of cDNA used. Reaction products were analyzed using 2% agarose gel electrophoresis to confirm that the signals detected by the GeneAmp PCR system 7700 (Perkin-Elmer Corporation, Foster City, USA) were from the expected products. Three independent experiments were performed.
Table II. Sequences of primers used for PCR.
| Gene | Sense | Antisense |
|---|---|---|
| β-Actin | AATCTGGCACCACACCTTCTAC | GCTTCTCCTTAATGTCACGCAC |
| GSTT1 | GCATAAGGTGATGTTCCCTGTGT | CGGTGCAAGGGTGAGGTTTC |
| ABCC2 | GACATCTATCTTCTAGATGACC | TAGATGGAGAACTTCACCTT |
| TOP2A | GGGTAGCAATAATCTAAACCTC | CCAGTTCTTCAATAGTACCCT |
| Caspase 3 | TGAAGCTACCTCAAACTTCC | CAGCATCACTGTAACTTGCT |
Results
Using CD-DST, valid T/C ratios were obtained in 9 (64.3%) of the 14 cancers. A T/C ratio of 50% or less was regarded as indicating that cells were gemcitabine-sensitive in vitro (Table III). On this basis, four cancers were classified as gemcitabine-sensitive and five as gemcitabine-resistant. However, an arbitrarily assigned growth inhibition rate may not always reflect clinical response because clinical response needs to be based on log killed cells.
Table III. The log2 transformed Cy3/Cy3 signals of selected genes.
| Sensitivity | Case no. | GSTT1 | ABCC2 | TOP2A | Caspase 3 |
|---|---|---|---|---|---|
| Sensitive | Case 1 | 0.18 | −0.24 | 1.10 | 0.30 |
| Case 11 | 1.25 | −1.50 | 1.37 | −1.25 | |
| Case 12 | −0.19 | 0.38 | −0.18 | 1.26 | |
| Case 14 | 0.16 | −0.22 | 0.19 | 0.09 | |
| Resistant | Case 3 | 1.20 | 0.29 | −0.25 | −2.54 |
| Case 4 | 2.80 | 1.20 | −2.33 | −1.92 | |
| Case 5 | 1.72 | 1.87 | −0.32 | −1.78 | |
| Case 6 | 2.05 | 1.07 | 0.11 | –0.52 | |
| Case 9 | 3.58 | 2.45 | −1.78 | −1.77 | |
| p value | <0.05 | <0.05 | <0.05 | <0.05 | |
Expressions of GSTT1, ABCC2, TOP2A, and caspase 3 between the gemcitabine-sensitive group and gemcitabine-resistant group are significantly different (p<0.05).
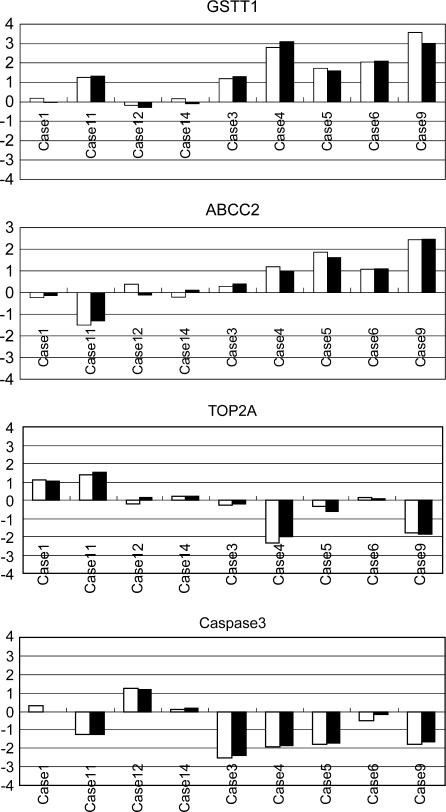
The log2 transformed Cy3/Cy3 signal data from microarray analyses are shown in Table III (original data are available to readers upon request by e-mail). In all 1081 human genes contained in the Atlas Glass Human 1.0 microarray, statistical analysis of the microarray data identified 4 genes that were differentially expressed between gemcitabine-sensitive and -resistant cancers: microsomal glutathione S-transferase 1 (GSTT1), topoisomerase II alpha (TOP2A), caspase 3, and ATP-binding cassette subfamily C member 2 (ABCC2). Real-time PCR analyses confirmed the differential expression of these genes (Figure 1). Paired Student's t test showed no difference between the results of the two methods (p>0.05). The fluctuations in mRNA expression between the nine patients were found to be similar using either analytical method.
Figure 1. .
Real-time PCR analyses (▪) for confirmation of microarray data (□). The same RNA source was used for both microarray and real-time PCR analyses. The Y-axis indicates the log2 transformed ratio of mRNA expression. Paired Student's t test showed no difference between the results of two methods (p>0.05).
For a further 22 genes, while statistical analysis indicated that the difference in their expression between the two tissues was not significant, the p values were close to 0.05. These genes were associated with gemcitabine resistance and included cyclin-dependent kinase inhibitor 1A, tumor protein p53 binding protein 2, activated p21cdc42Hs kinase, v-akt 2, insulin-like growth factor 1 receptor, BCL2-interacting killer, BCL2-like 2, BCL2-like 1, BCL2-related protein A1, BCL2-interacting killer, topoisomerase I, APEX nuclease, transforming growth factor beta receptor II, interleukin 6 receptor, cytochrome P450 subfamily 1 (dioxin-inducible), polypeptide 1, glutathione S-transferase M1, transforming growth factor beta 1, interleukin 8, insulin-like growth factor 1, nuclear factor of kappa light polypeptide gene enhancer in B-cells 2, ligase III and ligase I.
Discussion
Gemcitabine is a nucleoside analog which exhibits metabolic characteristics that distinguish it from related compounds and may explain its activity against solid tumors. The active nucleotide effectively accumulates at high concentrations in cells due to both efficient phosphorylation and relatively slow elimination. The diphosphate is a potent inhibitor of ribonucleotide reductase, resulting in reduced deoxynucleotide pools. Decreased cellular concentrations of deoxycytidine triphosphate permit more rapid phosphorylation of gemcitabine and decrease the metabolic clearance of gemcitabine nucleotides by deoxycytidine monophosphate deaminase. Most importantly, increasing the ratio of the cellular concentration of gemcitabine triphosphate to deoxycytidine triphosphate increases analog incorporation into DNA, which is strongly associated with loss of cell viability 7. Gemcitabine alone or in combination with other anticancer drugs has become a popular regimen in pancreatic cancer.
Analysis of gene expression using cDNA chips showed that four genes were differentially expressed according to cells being either gemcitabine-sensitive or -resistant. The four genes were identified as GSTT1, TOP2A, caspase 3, and ABCC2. For each gene, expression was associated with drug resistance. These findings are in agreement with a report by Scherf et al. indicating that gene expression profiles may reflect drug sensitivity in cancer cells 8.
The cellular glutathione system (GSH) is a critical component of the cytostatic detoxification pathway in cells. GSTT1 is a member of a protein superfamily that catalyzes the conjugation of reduced glutathione to a variety of electrophilic and hydrophobic compounds. The conjugate is less active and more water-soluble, and it is excluded from the cell with the participation of GS-X transporter proteins. GSTT1 is claimed to play an important role in human carcinogenesis 9,10. Inhibitors of glutathione transferases have been shown to enhance the cytotoxicity of alkylating chemotherapeutic drugs in cultured cancer cells otherwise resistant to this class of agent 11.
ABCC2 (MRP2) is a member of the ATP-binding cassette (ABC) transporter superfamily, which is involved in biliary, renal, and intestinal secretion of numerous organic anions, including endogenous compounds such as bilirubin and exogenous compounds such as drugs and toxic chemicals 12. This protein is a member of the MRP subfamily that is involved in multi-drug resistance 13.
TOP2A functions as the target for several anticancer agents, and a variety of mutations in this gene have been associated with drug resistance 14. The activity or quantity of this enzyme was lower in cell lines resistant to topoisomerase II-inhibiting drugs. In those lines, topoisomerase gene mutations were found which were presumed to be the bases for the drug resistance 15,16.
Caspase 3 cleaves and activates caspases 6, 7, and 9, is processed by caspases 8, 9, and 10, and plays an important role in apoptosis. A broad spectrum of anticancer drugs used in the clinic has been shown to activate apoptosis in vitro and in vivo17,18. Low expression of caspase 3 has been shown to inhibit chemotherapy-induced apoptosis 19.
In the present study, differences in the expression of a further 22 genes almost reached statistical significance. Many of these candidate genes have been previously linked to drug resistance or carcinogenesis. Some of the expressed sequence tags (ESTs) identified may represent genes that might be future targets for novel anticancer drugs. Identification of further differentially expressed genes will enable development of an accurate drug response system (DRS) for predicting the suitability of a particular cancer patient for gemcitabine therapy 20.
Conclusions
Microarray evaluation has a number of distinct advantages compared with the CD-DST method, such as the requirements for less tissue and less time (3 days), and the ease with which experiments can be repeated if required. These advantages are of particular importance in pancreatic cancer analysis where it is difficult to obtain large amounts of cancer tissue. We believe that clinical application of such a DRS will prevent cancer patients from undergoing ineffective adjuvant chemotherapy.
Acknowledgements
This work appeared in the Poster Prize Session at the 7th World Congress of the International Hepato-Pancreato-Biliary Association in Edinburgh, September 2006.
References
- 1.DeVita VT, Hellman S, Rosenberg SA, eds. Cancer: principles and practice of oncology, 6th edn. Philadelphia: Lippincott Williams and Wilkins, 2001:1126–61. [Google Scholar]
- 2.Do JK, Kubota T, Ura HT, Yamaue H, Akiyama S, Maehara Y, et al. Cumulative results of chemosensitivity tests for antitumor agents in Japan. Anticancer Res. 2000;20:2389–92. [PubMed] [Google Scholar]
- 3.Kobayshi H, Tanisaka K, Kondo N, Mito Y, Koezuka M, Yokouchi H, et al. Development of new in vitro chemosensitivity test using collagen gel droplet embedded culture and its clinical usefulness. Gan To Kagaku Ryoho 1995Nov;/22(13):/ 1933–9. Japanese. [PubMed] [Google Scholar]
- 4.Assersohn L, Gangi L, Zhao Y, Dowsett M, Simon R, Powles TJ, et al. The feasibility of using fine needle aspiration from primary breast cancer for cDNA microarray analyses. Clin Cancer Res. 2002;8:794–801. [PubMed] [Google Scholar]
- 5.Yasuda H, Takada T, Wada K, Amano H, Isaka T, Yoshida M, et al. A new in-vitro drug sensitivity test (collagen-gel droplet embedded-culture drug sensitivity test) in carcinomas of pancreas and biliary tract: possible clinical utility. J Hepatobiliary Pancreat Surg. 1998;5:261–8. doi: 10.1007/s005340050044. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi H. Collagen gel droplet culture method to examine in vitro chemosensitivity. Methods Mol Med 2005;/110:/59–67. [DOI] [PubMed] [Google Scholar]
- 7.Plunkett W, Huang P, Searcy CE, Gandhi V. Gemcitabine: preclinical pharmacology and mechanisms of action. Semin Oncol. 1996;23(5 Suppl 10):3–15. [PubMed] [Google Scholar]
- 8.Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–44. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 9.Trizna Z, Clayman GL, Spitz MR, Briggs KL, Goefert H. Glutathione s-transferase genotypes as risk factors for head and neck cancer. Am J Surg. 1995;170:499–501. doi: 10.1016/s0002-9610(99)80339-0. [DOI] [PubMed] [Google Scholar]
- 10.Strange RC, Lear JT, Fryer AA. Glutathione s-transferase polymorphisms: influence on susceptibility to cancer. Chem Biol Interact. 1998 Apr 24;111–112:351–64. doi: 10.1016/s0009-2797(97)00172-5. [DOI] [PubMed] [Google Scholar]
- 11.Awasthi S, Srivastava SK, Ahmad F, Ahmad H, Ansari GA. Interactions of glutathione s-transferase-pi with ethacrynic acid and its glutathione conjugate. Biochim Biophys Acta. 1993;1164:173–8. doi: 10.1016/0167-4838(93)90245-m. [DOI] [PubMed] [Google Scholar]
- 12.Payen L, Sparfel L, Courtois A, Vernhet L, Guillouzo A, Fardel O. The drug efflux pump MRP2: regulation of expression in physiopathological situations and by endogenous and exogenous compounds. Cell Biol Toxicol. 2002;18:221–3. doi: 10.1023/a:1016020626941. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki H, Sugiyama Y. Single nucleotide polymorphisms in multidrug resistance associated protein 2 (MRP2/ABCC2): its impact on drug disposition. Adv Drug Deliv Rev. 2002;54:1311–31. doi: 10.1016/s0169-409x(02)00075-3. [DOI] [PubMed] [Google Scholar]
- 14.Holden JK. Human deoxyribonucleic acid topoisomerase: molecular targets of anticancer drugs. Ann Clin Lab Sci. 1997;27:402–12. [PubMed] [Google Scholar]
- 15.Pommier Y, Leteurte F, Fesen MR, Fujimori A, Bertrand R, Solary E, et al. Cellular determinants of sensitivity and resistance to DNA topoisomerase inhibitors. Cancer Invest. 1994;12:530–42. doi: 10.3109/07357909409021413. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto Y, Takano H, Kunishio K, Nagao S, Fojo T. Hypophosphorylation of topoisomerase II alpha in etoposide (VP-16)-resistant human carcinoma cell lines associated with carboxy-terminal truncation. Jpn J Cancer Res. 2001;92:799–805. doi: 10.1111/j.1349-7006.2001.tb01164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dive C, Hickman JA. Drug-target interactions: only the first step in the commitment to a programmed cell death? Br J Cancer. 1991;64:192–6. doi: 10.1038/bjc.1991.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basu A, Castle VP, Bouziane M, Bhalla K, Haldar S. Crosstalk between extrinsic and intrinsic cell death pathways in pancreatic cancer: synergistic action of estrogen metabolite and ligands of death receptor family. Cancer Res. 2006;66:4309–318. doi: 10.1158/0008-5472.CAN-05-2657. [DOI] [PubMed] [Google Scholar]
- 19.Yang XH, Sladek TL, Liu X, Butler BR, Froelich CJ, Thor AD. Reconstitution of caspase sensitizes MCF-7 breast cancer cells to doxorubicin- and etoposide-induced apoptosis. Cancer Res. 2001;61:348–54. [PubMed] [Google Scholar]
- 20.Kihara C, Tsunoda T, Tanaka T, Yamana H, Furukawa Y, Ono K, et al. Prediction of sensitivity of esophageal tumors to adjuvant chemotherapy by cDNA microarray analysis of gene-expression profiles. Cancer Res. 2001;61:6474–9. [PubMed] [Google Scholar]