Abstract
Here, we disrupted the p70 S6 kinase (p70s6k) gene in murine embryonic stem cells to determine the role of this kinase in cell growth, protein synthesis, and rapamycin sensitivity. p70s6k−/− cells proliferated at a slower rate than parental cells, suggesting that p70s6k has a positive influence on cell proliferation but is not essential. In addition, rapamycin inhibited proliferation of p70s6k−/− cells, indicating that other events inhibited by the drug, independent of p70s6k, also are important for both cell proliferation and the action of rapamycin. In p70s6k−/− cells, which exhibited no ribosomal S6 phosphorylation, translation of mRNA encoding ribosomal proteins was not increased by serum nor specifically inhibited by rapamycin. In contrast, rapamycin inhibited phosphorylation of initiation factor 4E-binding protein 1 (4E-BP1), general mRNA translation, and overall protein synthesis in p70s6k−/− cells, indicating that these events proceed independently of p70s6k activity. This study localizes the function of p70s6k to ribosomal biogenesis by regulating ribosomal protein synthesis at the level of mRNA translation.
The serine/threonine kinase, p70s6k, is a unique kinase in higher eukaryotes and one whose function is believed to regulate protein synthesis (1). This kinase originally was identified in a mammalian cellular fraction to phosphorylate ribosomal S6 protein in vitro (2–4). p90rsk is another serine/threonine kinase that is also able to phosphorylate S6 in vitro. However, some extracellular stimuli such as insulin induce the activation of p70s6k and S6 phosphorylation without the activation of p90rsk (5, 6). Moreover, the macrolide rapamycin inhibits both p70s6k activity and S6 phosphorylation but not p90rsk activity, suggesting that p70s6k likely is responsible for in vivo S6 phosphorylation (7–11). Ribosomal S6 protein, located in a position in the 40S subunit, is thought to be a major functional site involved in tRNA and mRNA binding (12–14). The sites phosphorylated in vivo are clustered at the carboxyl terminus, and the phosphorylated region of S6 is thought to lie within the cleft where mRNA binds, a location compatible with a role in the regulation of translation (15). Indeed, S6 phosphorylation correlates with rates of mRNA translation (16). Taken together, p70s6k is a good candidate kinase for regulation of mRNA translation through S6 phosphorylation.
Rapamycin binds to FKBP12 in cells and inhibits the p70s6k activity through binding to a third molecule termed FRAP (or mTOR, RAFT) (17). Indeed, addition of rapamycin to cell cultures inhibits protein synthesis and, more specifically, translation of mRNAs encoding ribosomal proteins (18, 19), suggesting that p70s6k and S6 are important in these events. However, rapamycin also inhibits a variety of other cell growth-related events, including the activity of cyclin-dependent kinases (20, 21), phosphorylation of CREM (22), and phosphorylation of 4E-BP1 (23, 24). In particular, 4E-BP1 (also termed PHAS1) is another regulatory molecule for mRNA translation. The drug induces accumulation of the dephosphorylated species of 4E-BP1 that binds to a translation initiation factor eIF-4E and suppresses translation initiation of cap-dependent mRNAs. Thus, the effects of rapamycin on mRNA translation and protein synthesis may be exerted through its inhibition of 4E-BP1 phosphorylation. To this end, we disrupted the p70s6k gene in murine embryonic stem (ES) cells to define the function of p70s6k in protein synthesis and cell growth, and also to define the role of the kinase in rapamycin-sensitive events.
MATERIALS AND METHODS
Targeted Disruption of the p70s6k Gene in Mouse Embryonic Stem Cells.
A portion (14 kb) of the p70s6k gene was cloned from the 129SV mouse genomic library in the lambda FIXII vector (Stratagene) using full-length, 1.5-kb rat p70s6k cDNA. The neo selection cassette was inserted in the opposite orientation at BamHI and ApaI sites, to disrupt an exon encoding a part of the catalytic domain (corresponding amino acids 207–237 in the p70s6k protein). The downstream coding region is designed to be frame-shifted. PGKtk cassette (25) was then cloned at the 3′ SpeI site. The resulting targeting vector featured 5 kb of homologous sequence on the long arm and 1.2 kb on the short arm. The R1 ES cell line was cultured in DMEM (GIBCO) supplemented with 15% fetal calf serum (FCS) and leukemia inhibitory factor (1,000 units/ml, GIBCO) and transfected with the targeting vector as described (26). Colonies were picked after 12 days in a selection medium containing 0.5 mg/ml G418 (GIBCO) and 2 μM gancyclovir (Syntex, Palo Alto, CA) (26). Homologous recombination of the gene was screened by PCR and confirmed by Southern blotting. For isolating cells homozygous for the targeted allele, a heterozygous clone was grown in 6 mg/ml G418 (27). Resistant colonies were picked after 10 days in the high-concentration selection medium.
Southern Blotting.
Genomic DNA was extracted from cultured cells, digested with PstI and EcoRI, and separated in 1% agarose gels. After denaturation and neutralization of the gel, DNA was transferred to nylon membranes and hybridized with a specific probe.
Western Blotting.
Cells (5 × 106) were washed with PBS and lysed at 4°C with 25 mM Tris⋅HCl, pH 7.4/50 mM NaCl/0.5% sodium deoxycholate/2% Nonidet P-40 (NP40)/0.2% SDS/1 μM phenylmethylsulfonyl fluoride (PMSF)/50 μg/ml aprotinin/50 μM leupeptin. Lysates were resolved by SDS/8% (for p70/85s6k and p90rsk) or 15% (for 4E-BP1) polyacrylamide gels and transferred to nitrocellulose filters. After blocking of the filters with a solution containing 1% BSA, the filters were incubated with either a rabbit polyclonal antibody raised against the common carboxyl-terminal sequence (NSGPYKKQAFPMISKRPEHLRMNL) of the p70s6k and p85s6k isoforms (C18, Santa Cruz Biotechnology), a rabbit polyclonal antibody raised against a 44-aa peptide representing residues 682–724 of p90rsk (QLVKGAMAATYSALNSSKPTPQLKPIESSILAQRRVRKLPSTTL) (anti-mouse rsk kinase, rsk-III, Upstate Biotechnology, Lake Placid, NY), or a rabbit polyclonal antibody raised against a recombinant His-tagged rat PHAS1 (4E-BP1), kindly provided by R. T. Abraham (Mayo Clinic, Rochester, MN) (28). Specific reactive proteins were detected by an enhanced chemiluminescence (ECL) method, employing a donkey anti-rabbit Ig antibody linked to horseradish peroxidase (Amersham).
Kinase Activity.
The specific activities of p70/85s6k were determined by [32P] incorporation into S6 peptide in the immune complex as described previously (29). Briefly, cells (5 × 106) were washed with PBS and lysed at 4°C in 500 μl of lysis buffer (10 mM potassium phosphate/1 mM EDTA/5 mM EGTA/10 mM MgCl2/50 mM β-glycerophosphate/1 mM Na3VO4/2 mM DTT/40 μg/ml PMSF/0.1% NP40). The extract was incubated for 30 min at 4°C with the C18 antibody. The immune complex was absorbed to Protein G-coupled beads (Zymed) for 30 min and washed twice with the lysis buffer and once with kinase buffer (20 mM Tris⋅HCl, pH 7.5/10 mM MgCl2/1 μg/ml IP-20/0.1 mg/ml BSA/0.4 mM DTT). After the final wash, the immune complexes were suspended in 50 μl of the kinase buffer containing 100 μM unlabeled ATP, 200 μCi/ml [γ-32P]ATP (1 Ci = 37 GBq), and 125 μM S6 peptide (RRRLSSLRA, Upstate Biotechnology). The reaction was allowed to proceed for 15 min at 30°C and terminated by the addition of 20 μl of 250 mM EDTA and boiling for 5 min. After a brief centrifugation, the supernatant (25 μl) was applied to phosphocellulose paper and radioactivity was determined by using a liquid scintillation counter.
S6 Phosphorylation in Vivo.
Cells were incubated with 500 μCi/ml of [32P]orthophosphate (ICN) in phosphate-free medium for 3 hr in the presence or absence of rapamycin. Cells were lysed in a hypotonic buffer, and ribosomes were enriched by ultra centrifugation using a sucrose cushion as described previously (6, 30). Ribosomal proteins then were separated on SDS/10% polyacrylamide gels, and phosphorylated S6 (≈32 kDa) was visualized by using the PhosphorImager and image quant analysis (Molecular Dynamics).
Polysomal Association.
Polysomal mRNAs were separated from nonpolysomal mRNAs by sucrose gradient gels as described previously (19). Briefly, cells (2 × 107) were washed once with PBS, they were suspended for 10 min at 0°C in 1 ml of a buffer containing 10 mM NaCl, 10 mM Tris⋅HCl (pH 7.4), 15 mM MgCl2, 1.2% Triton X-100, 1.2% deoxycholate, and a ribonuclease inhibitor, RNasin 200 units/ml (Promega). Nuclei were pelleted by centrifugation for 2.5 min at 10,000 × g. The postnuclear supernatant (300 μl) was layered over 11 ml of a 0.5–1.5 M sucrose gradient with a 1-ml cushion of 1.5 M sucrose. The sucrose solutions contained 2 mM Tris⋅HCl (pH 7.4), 25 mM NaCl, 5 mM MgCl2, and 100 μg of heparin per ml. The gradients were centrifuged at 105,000 × g for 3 hr at 4°C by using a Beckman SW28 rotor. Twelve fractions of 1 ml each were collected and RNA was extracted. RNA from each fraction then was applied to nitrocellulose membranes by using a slot blot apparatus. Membranes were hybridized with 32P-labeled probes of human eEF-1α cDNA, hamster eEF-2 cDNA, human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA, and human β-actin cDNA (ref. 19; ref. 29 and references therein). Radioactivity of slot blots was scanned by using the PhosphorImager, and the relative counts in each fraction were determined by image quant analysis.
RESULTS
Targeted Disruption of the p70s6k Gene.
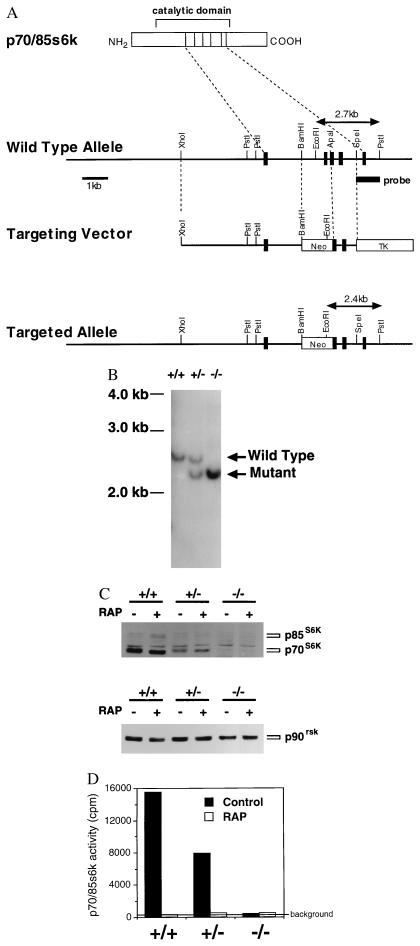
We generated mouse ES cells lacking p70s6k expression by using homologous recombination (Fig. 1A). Mutation of both p70s6k alleles (p70s6k−/−) was established from p70s6k+/− ES cells by selection in high doses of G418 (Fig. 1B). p70s6k protein was absent in p70s6k−/− ES cells (Fig. 1C). As a control, comparable protein levels of p90rsk in all of the samples are shown (Fig. 1C). It should be noted that p70s6k has a p85s6k isoform that has 23 additional amino acids at its amino terminus (31–33). The p70s6k and p85s6k isoforms are produced by alternative splicing of the first exons (31). As is predicted by the fact that we disrupted the common exons of p70s6k and p85s6k encoding the catalytic domain (Fig. 1A), the p85s6k isoform also was absent in p70s6k−/− ES cells (Fig. 1C). We confirmed that there was no activity of p70/85s6k detected in the p70s6k−/− ES cells by using antibodies raised against the common carboxyl-terminal sequence (Fig. 1D).
Figure 1.
Targeted disruption of the p70s6k gene in mouse ES cells. (A) Structure of the wild-type allele, targeting vector, and targeted allele. A portion (14 kb) of the p70/85s6k gene was cloned from the 129SV mouse genomic library. The neo selection cassette was inserted in the opposite orientation at BamHI and ApaI sites to disrupt an exon encoding a part of the catalytic domain (corresponding amino acids 207–237 in the p70s6k protein). The downstream coding region is designed to be frame-shifted. PGKtk cassette was then cloned at the 3′ SpeI site. The resulting targeting vector featured 5 kb of homologous sequence on the long arm and 1.2 kb on the short arm. (B) Southern blotting. The R1 ES cell line was transfected with the targeting vector. Colonies were picked after 12 days in selection medium and expanded. For isolating cells homozygous for the targeted allele, a heterozygous clone was grown on feeder cells in 6 mg/ml G418. Resistant colonies were picked after 10 days in high-concentration selection medium. Genomic DNA was extracted from cultured cells, digested with PstI and EcoRI, and separated in 1% agarose gel. After denature and neutralization of the gel, DNA was transferred to nylon membrane and hybridized with the 1.0-kb probe indicated in A. (C) Western blotting. Cells were treated with either vehicle (0.1% ethanol, Control) or rapamycin (10 ng/ml, RAP) for 30 min. Proteins were extracted from cells, separated on SDS/8% polyacrylamide gels, and transferred to nitrocellulose membranes. Immunoblotting was performed by using the ECL method. Antibodies used here were: for p70s6k and p85s6k, a rabbit polyclonal antibody raised against the common carboxyl-terminal sequence (C18); for p90rsk, a rabbit polyclonal antibody raised against the carboxyl terminus of the kinase. (D) Kinase activity. Cells were prepared as described above. The specific activities of p70s6k and p85s6k were measured by using the C18 antibody and S6 peptide as a substrate.
Proliferation of p70s6k−/− Cells.
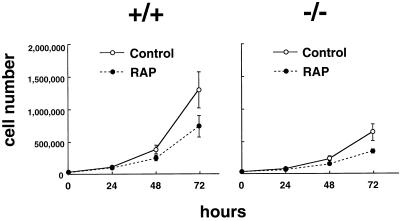
p70s6k is activated by various mitogenic stimuli, including epidermal growth factor, platelet-derived growth factor, insulin, and interleukin 2, and the activity remains high throughout the G1 phase of the cell cycle (34, 35). Kinase activation is thought to be essential for G1/S transition because microinjection of specific antibodies against p70s6k abolishes the entry of the cells into the cell cycle (36). Contrary to this previous observation, p70s6k−/− cells proliferated well (Fig. 2), indicating that p70s6k is not essential for G1/S transition. However, p70s6k−/− cells proliferated slower than parental cells (Fig. 2), whereas p70s6k+/− cells demonstrated a similar growth rate as parental cells (data not shown). These data were reproducible over five independent experiments, and the slower-growing phenotype of p70s6k−/− cells was observed even when p70s6k−/− cells were replated in 2- to 5-fold higher densities than parental cells (data not shown). Cell cycle analysis demonstrated that 33.5 ± 2.9% of the p70s6k+/+ cells are within the G0/G1 stage with 2 N content of DNA, whereas 53.1 ± 2.3% of p70s6k−/− cells are in G0/G1 (mean ± SD from three independent experiments using cells harvested 1 and 2 days after passage). These results indicate that p70s6k activity has a positive influence on cell proliferation, especially on G1 progression. Of interest, rapamycin inhibited cell proliferation of p70s6k−/− cells to a similar extent as parental cells, indicating that other events inhibited by the drug but independent of p70s6k are also important for both cell proliferation and the action of rapamycin. Rapamycin is known to inhibit the activity of cyclin-dependent kinases (Cdks) through inhibition of elimination of p27kip1 under certain conditions (20, 21). Here, Cdk2 activity was not inhibited by a 3-hr treatment with rapamycin and was inhibited by approximately 30% after 24-hr treatment with rapamycin in both p70s6k+/+ and p70s6k−/− cells. These data suggest that Cdk2 activity is not a primary target of either rapamycin or p70s6k, at least in embryonic stem cells.
Figure 2.
Cell proliferation. p70s6k+/+ ES cells or p70s6k−/− ES cells were incubated in DMEM medium supplemented with 15% FCS and leukemia inhibitory factor (1,000 units/ml), in the presence or absence of rapamycin (10 ng/ml) for 72 hr. Cells were counted by using a Coulter counter (Hialeah, FL).
In Vivo Phosphorylation of S6 and 4E-BP1.
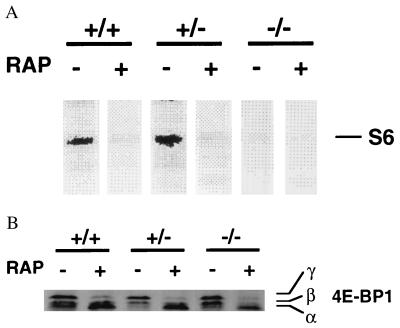
Ribosomal S6 protein is the best candidate for the in vivo substrate of p70s6k. The cells were metabolically labeled with [32P]orthophosphate in the presence or absence of rapamycin, and radiolabeled proteins in partially purified ribosomes were separated on SDS/polyacrylamide gels (Fig. 3A). In contrast to p70s6k+/+ cells or p70s6k+/− cells, which exhibited rapamycin-sensitive S6 phosphorylation, p70s6k−/− cells did not show any detectable S6 phosphorylation in vivo. The data establish that p70s6k is the in vivo kinase for S6 phosphorylation.
Figure 3.
In vivo phosphorylation of S6 and 4E-BP1. (A) S6 phosphorylation in vivo. Cells were incubated with [32P]orthophosphate in phosphate-free medium in the presence or absence of rapamycin (10 ng/ml) for 3 hr. Cells were lysed in hypotonic buffer, and ribosomes were enriched by ultra centrifugation using a sucrose cushion. Ribosomal proteins were then separated on SDS/10% polyacrylamide gels, and phosphorylated S6 (≈32 kDa) was visualized by using the PhosphorImager and image quant analysis (Molecular Dynamics). (B) In vivo phosphorylation status of 4E-BP1. Cells were treated with vehicle or rapamycin as described in Fig. 1C. Protein extracts were separated on SDS/15% polyacrylamide gels and immunoblotted by using a rabbit polyclonal antibody raised against 4E-BP1. The band with the highest mobility (α) corresponds to hypophosphorylated 4E-BP1, and the bands with lower mobility (β, γ) correspond to hyperphosphorylated 4E-BP1 (23, 24, 28).
The phosphorylation status of 4E-BP1 also is altered by rapamycin (38, 39). The drug induces accumulation of the dephosphorylated species of 4E-BP1, as well as p70s6k and S6. The in vivo phosphorylation status of 4E-BP1 was examined by its mobility in SDS/polyacrylamide gels as described previously (23, 24, 28). In contrast to S6, phosphorylation of 4E-BP1 was detected in p70s6k−/− cells to the same extent as in parental cells (Fig. 3B). Furthermore, rapamycin induced the accumulation of a dephosphorylated form of 4E-BP1 in p70s6k−/− cells. These data indicate that 4E-BP1 phosphorylation is independent of p70s6k activity. This is consistent with recent publications demonstrating that 4E-BP1 is a direct substrate of FRAP (28) or that 4E-BP1 phosphorylation is controlled by a parallel signaling pathway that bifurcates immediately upstream of p70s6k (37).
Translation of mRNA Encoding Ribosomal Proteins.
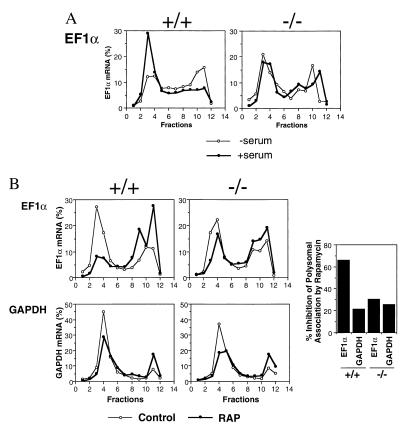
We and others previously reported that rapamycin selectively inhibits translation of mRNAs that encode ribosomal proteins and elongation factors (18, 19). This set of mRNAs has a specific structure at the 5′ terminus (termed 5′TOP mRNA for mRNAs with an oligopyrimidine tract at their transcriptional start). The transcription initiates at cytidine and is followed by at least a 6-pyrimidine stretch. This is usually followed by GC-rich regions in the 5′ untranslated region, which potentially make loop–stem structures. More recently, it has been demonstrated that the activity of p70s6k correlates with the translational status of 5′TOP mRNAs by using dominant-negative or rapamycin-resistant forms of p70s6k (38). We examined the status of polysomal association of elongation factor 1α (EF-1α) mRNA, as a representative of the 5′TOP group of mRNAs, and of GAPDH mRNA as a representative of non-5′TOP mRNAs (Fig. 4). Addition of serum after a 16-hr period of starvation increased the polysomal-associated fractions (fractions 2–6) of EF-1α mRNA in p70s6k+/+ cells, but not p70s6k−/− cells (Fig. 4A), indicating that p70s6k is responsible for the up-regulation of 5′TOP mRNA translation by mitogens. In contrast, addition of serum did not change the polysomal association of GAPDH mRNA in either p70s6k+/+ or p70s6k−/− cells (data not shown). It should be noted that about 40% of EF-1α mRNA was detected in the polysomal fractions of p70s6k−/− cells, regardless of the concentrations of serum. This result was intermediate between serum-starved p70s6k+/+ cells and serum-added p70s6k+/+ cells. Currently, it is unclear why p70s6k−/− cells exhibit this intermediate value of 5′TOP mRNA translation. A compensatory mechanism downstream of p70s6k/S6 might partially up-regulate 5′TOP mRNA in p70s6k−/− cells. This possibility includes the decrease in a repressor protein that binds specifically to 5′TOP mRNA.
Figure 4.
Polysomal association of EF-1α mRNA and GAPDH mRNA. (A) Effects of serum deprivation/addition. Cells were incubated in medium containing 0.1% FCS for 16 hr (−serum). FCS (15%) was added, and cells were incubated for another 3 hr (+serum). Polysomal mRNAs were separated from nonpolysomal mRNAs by sucrose gradient gels. mRNA in each fraction was slot-blotted and hybridized with a 32P-labeled cDNA probe for human EF-1α. Radioactivity of slot blots was scanned by using the PhosphorImager, and the relative counts in each fraction were determined by image quant analysis (shown as a percentage of the total counts in 12 fractions). Fractions 2–6 correspond to polysomal fractions determined by OD260 measurement (data not shown). (B) Effects of rapamycin. Cells were incubated in medium containing 0.1% FCS for 16 hr, then incubated in medium containing 15% FCS (Control) or 15% FCS plus rapamycin (10 ng/ml, RAP) for an additional 3 hr. Polysomal association of EF-1α mRNA or GAPDH mRNA was measured as described above. The percent inhibition of polysomal-associated EF-1α mRNA or GAPDH mRNA by rapamycin also is shown (Right).
The effects of rapamycin then were investigated in the p70s6k−/− cells (Fig. 4B). In p70s6k+/+ cells, rapamycin profoundly reduced the polysomal association of EF-1α mRNA as we previously reported in other cell types (19). Rapamycin also reduced the polysomal association of GAPDH mRNA, but to a much lower extent when compared with EF-1α mRNA (Fig. 4B Right). This lower degree of inhibition in non-5′TOP mRNA association may be explained by the general inhibition of translational initiation through the inhibition of eIF-4E activity. Essentially identical results were obtained in p70s6k+/− cells (data not shown). In contrast, the specific effects of rapamycin on 5′TOP mRNA were eliminated in p70s6k−/− cells. Rapamycin inhibited polysomal association of both EF-1α mRNA and GAPDH mRNA to the same extent as it did for GAPDH mRNA in p70s6k+/+ cells. These findings were reproduced for another set of 5′TOP mRNA (EF-2 mRNA) and non-5′TOP mRNA (β-actin mRNA) (data not shown). These results indicate that rapamycin exerts its particular inhibitory effects on 5′TOP mRNA translation through p70s6k. The data also indicate that inhibition of more general translation is independent of p70s6k, consistent with the data above showing that rapamycin inhibited 4E-BP1 phosphorylation in p70s6k−/− cells as well. The inhibition of overall protein synthesis in p70s6k+/+ cells and p70s6k−/− cells also was examined by incorporation of [3H]amino acids into cells. Treatment of the cells with rapamycin for 3 hr inhibited protein synthesis by 34.2 ± 6.3% and 26.1 ± 3.7% (mean ± SD, from six independent experiments) in p70s6k+/+ cells and p70s6k−/− cells, respectively. These data indicate that inhibition of overall protein synthesis by rapamycin is independent of p70s6k activity for the most part. In addition, serum-induced, rapamycin-resistant p90rsk activation similarly was observed in both p70s6k+/+ cells and p70s6k−/− cells, indicating that the absence of p70s6k does not have an obvious effect on early events in mitogenic signaling (data now shown).
DISCUSSION
Deletion of the p70s6k gene was achieved in ES cells by homologous recombination to define the function of p70s6k in mammalian cells. Targeted disruption of the p70s6k gene eliminated phosphorylation of ribosomal S6 protein and abrogated translational regulation of mRNAs encoding ribosomal proteins by serum and rapamycin in these murine ES cells. These data demonstrate that the function of p70s6k resides in S6 phosphorylation and also in regulation of ribosomal protein synthesis at the level of mRNA translation. Ribosomal proteins are needed in equimolar amounts in ribosomal biogenesis. In prokaryotes, genes for ribosomal proteins are interspersed in a few operons and the translation of these operons are autogenously regulated by one of their products (39). The translation of an operon is down-regulated whenever one of its products (i.e., a ribosomal protein) is in excess compared with the amounts of rRNA. Each of the regulators is a ribosomal protein that binds directly to rRNA and also to its own operon mRNA (40). It is not likely that the synthesis of eukaryotic ribosomal proteins is regulated by a similar feedback mechanism, based on studies introducing extra copies of ribosomal protein genes or mRNAs into cells [see discussion in Hammond and Bowman (ref. 41 and references therein)]. The present study indicates that phosphorylation, achieved by the serine/threonine kinase p70s6k, is an alternative mechanism for coordinate regulation of ribosomal protein synthesis in mammalian cells. In addition, phosphorylation of a ribosomal protein itself (S6) is a candidate for this regulation. 5′TOP mRNA (or 5′TOP mRNA bound to a specific repressor protein) may have a higher affinity to the small subunit of ribosome (40S) containing phosphorylated S6 than to the subunit with unphosphorylated S6.
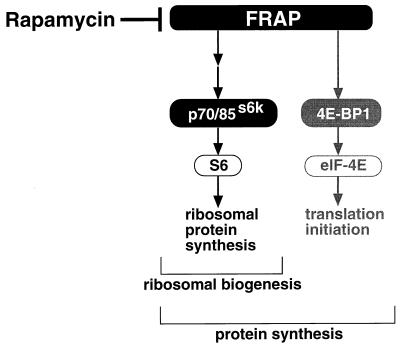
We also demonstrated that rapamycin inhibited 4E-BP1 phosphorylation, translation of general mRNA, and overall protein synthesis, independent of p70s6k activity. The hypophosphorylated species of 4E-BP1 bind tightly to eIF-4E (an N7-methylguanosine cap-binding subunit of the eIF-4F complex) and prevent eIF-4E from associating with eIF-4G (a scaffolding protein in the eIF-4F complex). The phosphorylation of 4E-BP1 is thought to release eIF-4E and facilitate translational initiation of capped mRNA. Thus, the inhibition of 4E-BP1 phosphorylation by rapamycin provides an explanation for the inhibition of overall mRNA translation and protein synthesis in p70s6k−/− cells. Our data distinguish the role of p70s6k from that of 4E-BP1 in the rapamycin-sensitive and FRAP-regulated pathway (Fig. 5). FRAP plays a pivotal role in the regulation of protein synthesis in cells by regulating the overall translation-initiation rate through 4E-BP1 and by regulating ribosomal biogenesis through p70s6k.
Figure 5.
A proposed model for the roles of p70s6k in protein synthesis.
Several aspects of the role of p70s6k remain to be clarified. For example, despite using several commercially available antibodies, we have not been able to analyze CREM phosphorylation to date. In terms of ribosomal biogenesis, rRNA also is required in the same equimolar amounts as ribosomal proteins. The same FRAP pathway or p70/85s6k by itself might regulate rRNA synthesis. Indeed, there is a report demonstrating that rapamycin at a high concentration (1 μg/ml) inhibits rRNA synthesis (42). The establishment of p70s6k−/− cells now permits direct investigation of the role of p70/85s6k in rRNA synthesis. Individual reconstitution of the p70s6k or p85s6k isoforms in p70s6k(−/−) ES cells also may delineate the differential roles of the two isoforms of the kinase.
Acknowledgments
This work was supported by Grants CA-64685 (N.T.) and HL-36577 (E.W.G.) from the National Institutes of Health. Authors are indebted to Dr. Robert T. Abraham (Mayo Clinic, Rochester, MN) for providing the antibody against 4E-BP1 (PHAS-1), Dr. Hitoshi Sasai (Japan Tobacco Inc., Yokohama, Japan), and Dr. James J. Lee (Mayo Clinic, Scottsdale, AZ) for providing materials and helpful suggestions for gene targeting.
ABBREVIATIONS
- p70s6k
p70 S6 kinase
- 4E-BP1
initiation factor 4E-binding protein 1
- ES cells
embryonic stem cells
- 5′TOP mRNA
mRNAs with an oligopyrimidine tract at their transcriptional start
- FCS
fetal calf serum
- EF-1α
elongation factor 1α
References
- 1.Brown E J, Schreiber S L. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 2.Blenis J, Kuo C J, Erikson R L. J Biol Chem. 1987;262:14373–14376. [PubMed] [Google Scholar]
- 3.Jeno P, Ballou L M, Novak-Hofer I, Thomas G. Proc Natl Acad Sci USA. 1988;85:406–410. doi: 10.1073/pnas.85.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price D J, Nemenoff R A, Avruch J. J Biol Chem. 1989;264:13825–13833. [PubMed] [Google Scholar]
- 5.Ballou L M, Luther H, Thomas G. Nature (London) 1991;349:348–350. doi: 10.1038/349348a0. [DOI] [PubMed] [Google Scholar]
- 6.Blenis J, Chung J, Erikson E, Alcorta D A, Erikson R L. Cell Growth Differ. 1991;2:279–285. [PubMed] [Google Scholar]
- 7.Calvo V, Crews C M, Vik T A, Bierer B E. Proc Natl Acad Sci USA. 1992;89:7571–7575. doi: 10.1073/pnas.89.16.7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung J, Kuo C J, Crabtree G R, Blenis J. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 9.Kuo C J, Chung J, Fiorentino D F, Flanagan W M, Blenis J, Crabtree G R. Nature (London) 1992;358:70–73. doi: 10.1038/358070a0. [DOI] [PubMed] [Google Scholar]
- 10.Price D J, Grove J R, Calvo V, Avruch J, Bierer B E. Science. 1992;257:973–977. doi: 10.1126/science.1380182. [DOI] [PubMed] [Google Scholar]
- 11.Terada N, Lucas J J, Szepesi A, Franklin R A, Takase K, Gelfand E W. Biochem Biophys Res Commun. 1992;186:1315–1321. doi: 10.1016/s0006-291x(05)81549-9. [DOI] [PubMed] [Google Scholar]
- 12.Terao K, Ogata K. J Biochem. 1979;86:597–603. doi: 10.1093/oxfordjournals.jbchem.a132563. [DOI] [PubMed] [Google Scholar]
- 13.Bommer U-A, Noll F, Lutsch G, Bielka H. FEBS Lett. 1980;111:171–174. doi: 10.1016/0014-5793(80)80785-x. [DOI] [PubMed] [Google Scholar]
- 14.Tolan D R, Traut R R. J Biol Chem. 1981;256:10129–10136. [PubMed] [Google Scholar]
- 15.Kozma S C, Ferrari S, Thomas G. Cell Signalling. 1989;3:219–225. doi: 10.1016/0898-6568(89)90039-9. [DOI] [PubMed] [Google Scholar]
- 16.Proud C G. Curr Top Cell Regul. 1992;32:243–369. doi: 10.1016/b978-0-12-152832-4.50008-2. [DOI] [PubMed] [Google Scholar]
- 17.Brown E J, Beal P A, Keith C T, Chen J, Shin T B, Schreiber S L. Nature (London) 1996;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 18.Jefferies H B J, Reinhard C, Kozma S C, Thomas G. Proc Natl Acad Sci USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terada N, Patel H R, Takase K, Kohno K, Nairn A C, Gelfand E W. Proc Natl Acad Sci USA. 1994;91:11477–11481. doi: 10.1073/pnas.91.24.11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nourse J, Firpo E, Flanagan W M, Coats S, Polyak K, Lee M-H, Massague J, Crabtree G R, Roberts J M. Nature (London) 1994;372:570–573. doi: 10.1038/372570a0. [DOI] [PubMed] [Google Scholar]
- 21.Kato J, Matsuoka M, Polyak K, Massague J, Sherr C J. Cell. 1994;79:487–496. doi: 10.1016/0092-8674(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 22.de Groot R P, Ballou L M, Sassone-Corsi P. Cell. 1994;79:81–91. doi: 10.1016/0092-8674(94)90402-2. [DOI] [PubMed] [Google Scholar]
- 23.Beretta L, Gingras A-C, Svitkin Y V, Hall M N, Sonenberg N. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 24.von Manteuffel S R, Gingras A-C, Ming X-F, Sonenberg N, Thomas G. Proc Natl Acad Sci USA. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudnicki M A, Braun T, Hinuma S, Jaenisch R. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 26.Hogan B. Manipulating the Mouse Embryo. New York: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 27.Mortensen R M, Conner D A, Chao S, Geisterfer-Lowrance A A T, Seidman J G. Mol Cell Biol. 1992;12:2391–2395. doi: 10.1128/mcb.12.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunn G J, Hudson C C, Sekulic A, Williams J M, Hosoi H, Houghton P J, Lawrence J C, Jr, Abraham R T. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 29.Terada N, Takase K, Papst P, Nairn A C, Gelfand E W. J Immunol. 1995;155:3418–3426. [PubMed] [Google Scholar]
- 30.Patel H R, Terada N, Gelfand E W. Biochem Biophys Res Commun. 1996;227:507–512. doi: 10.1006/bbrc.1996.1537. [DOI] [PubMed] [Google Scholar]
- 31.Grove J R, Banerjee P, Balasubramanyam A, Coffer P J, Price D J, Avruch J, Woodgett J R. Mol Cell Biol. 1991;11:5541–5550. doi: 10.1128/mcb.11.11.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinhard C, Thomas G, Kozma S C. Proc Natl Acad Sci USA. 1992;89:4052–4056. doi: 10.1073/pnas.89.9.4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinhard C, Fernandez A, Lamb N J C, Thomas G. EMBO J. 1994;13:1557–1565. doi: 10.1002/j.1460-2075.1994.tb06418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Susa M, Olivier A R, Fabbro D, Thomas G. Cell. 1989;57:817–824. doi: 10.1016/0092-8674(89)90796-4. [DOI] [PubMed] [Google Scholar]
- 35.Terada N, Franklin R A, Lucas J J, Blenis J, Gelfand E W. J Biol Chem. 1993;268:12062–12068. [PubMed] [Google Scholar]
- 36.Lane H A, Fernandez A, Lamb N J C, Thomas G. Nature (London) 1993;363:170–172. doi: 10.1038/363170a0. [DOI] [PubMed] [Google Scholar]
- 37.von Manteuffel S R, Dennis P B, Pullen N, Gingras A-C, Sonenberg N, Thomas G. Mol Cell Biol. 1997;17:5426–5436. doi: 10.1128/mcb.17.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jefferies H B J, Fumagalli S, Dennis P B, Reinhard C, Pearson R B, Thomas G. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewin B. Genes V. Oxford: Oxford Univ. Press; 1994. [Google Scholar]
- 40.Baughman G, Nomura M. Cell. 1983;34:979–988. doi: 10.1016/0092-8674(83)90555-x. [DOI] [PubMed] [Google Scholar]
- 41.Hammond M L, Bowman L H. J Biol Chem. 1988;263:17785–17791. [PubMed] [Google Scholar]
- 42.Mahajan P B. Int J Immunopharmacol. 1994;16:711–721. doi: 10.1016/0192-0561(94)90091-4. [DOI] [PubMed] [Google Scholar]