Abstract
Insect phenoloxidase (PO) participates in melanotic encapsulation, wound healing, and cuticle sclerotization. It is converted from prophenoloxidase (proPO) by a proPO-activating proteinase (PAP). Manduca sexta PAP-1, the final component of a serine proteinase cascade, cleaves proPO to generate active PO. In an effort to understand the transcriptional regulation, we isolated a genomic clone of the PAP-1 gene, determined its nucleotide sequence, and elucidated its exon–intron organization. Computer analysis revealed several immune and hormone responsive elements in the upstream region. Southern blot analysis suggested that the M. sexta genome contains a single copy of PAP-1 gene. Reverse transcription-polymerase chain reaction showed that PAP-1 was constitutively expressed in fat body, trachea, and nerve tissue of the fifth instar larvae. The mRNA levels in hemocytes and fat body markedly increased in response to a bacterial challenge. We also observed tissue-specific and developmental regulation of the gene’s transcription. Treating M. sexta fat body culture with 20-hydroxyecdysone reduced the PAP-1 mRNA level. These data indicated that the expression of PAP-1 gene is under the dual control of immune and hormonal signals.
Keywords: Phenoloxidase, Melanization, Hemolymph protein, Serine proteinase cascade, Insect immunity, Gene regulation, Tobacco hornworm, Ecdysone
1. Introduction
Insect phenoloxidase (PO) hydroxylates tyrosine to dopa and oxidizes dopa to dopaquinone. Quinones can undergo cyclization, oxidation, and polymerization to generate eumelanin (Ashida and Brey, 1998). They may also crosslink proteins and polysaccharides to form hardened cuticle, seal off wounds, and encapsulate invading parasites. Reactive oxygen intermediates produced during the process can be cytotoxic to microbes as well as host tissues/cells (Nappi and Vass, 2001). Therefore, insect PO is produced as an inactive zymogen prophenoloxidase (proPO), which requires proPO-activating proteinase (PAP) for proteolytic activation. PAP resides at the end of a largely unknown serine proteinase (SP) pathway triggered upon recognition of microbial cell surface molecules, such as lipopolysaccharide, peptidoglycan, and β-1,3-glucan (Ashida and Brey, 1998). SP inhibitors in the hemolymph regulate this enzyme system (Kanost, 1999).
PAPs have been purified and cloned from Bombyx mori (Satoh et al., 1999), Holotrichia diomphalia (Lee et al., 1998), Manduca sexta (Jiang et al., 1998, 2003a, 2003b), and Pacifastacus leniusculus (Wang et al., 2001). There is at least one regulatory clip domain at the amino terminus of these enzymes. Clip domains, initially identified in the horseshoe crab proclotting enzyme (Muta et al., 1990), are frequently found in arthropod SPs and serine proteinase homologs (SPHs) (Jiang and Kanost, 2000; Huang et al., 2000; Ross et al., 2003; Christophides et al., 2002). We have purified three PAPs from M. sexta prepupae, which cleave proPO at Arg51 and require two clip-domain SPHs to generate active PO. Like proPO, these clip-domain SPs and SPHs are produced in a precursor form and need cleavage for activation. While the proPO activation system is beginning to be understood at the protein level, little is known about transcriptional regulation of the cascade components.
The interactions between cis-regulatory elements and transcription factors determine the expressions of insect immune genes (Harshman and James, 1998; Engstrom, 1998). Cecropia immune-responsive factor was first identified in nuclear extracts from the Hyalophora cecropia pupae challenged with bacteria (Sun and Faye, 1992). Three Rel-family transcription factors (Dorsal, Dif, and Relish) were identified from Drosophila melanogaster (Engstrom, 1998). Similar to NF-κB in mammals, these proteins may form homodimers or heterodimers in response to different stimuli and control the differential transcription of immune genes (Hoffmann, 2003). Dorsal and Dif belong to the Toll pathway, whereas Relish is a member of the Imd pathway. NF-κB responsive elements have been identified at the 5′ end of antimicrobial peptide genes from many insects (Harshman and James, 1998; Engstrom, 1998). In Drosophila, the consensus sequence of these elements is GGGRAYYYYY (Hultmark, 1993). A similar responsive element has been identified upstream of M. sexta hemolin and lysozyme genes (Wang et al., 1995; Mulnix and Dunn, 1994), indicating that NF-κB-like proteins may also exist in lepidopteran insects to regulate immune gene expression. Recently, an enhancer was identified in an intron of the H. cecropia hemolin gene, which activates a reporter gene through Drosophila Dif (Roxstrom-Lindquist et al., 2002). Nevertheless, no NF-κB-like transcription activator has been documented in M. sexta so far.
GATA boxes are cis-regulatory elements commonly found at the 5′ end of insect immune genes. Their consensus sequence, WGATAA, was first identified in the chorion gene promoter region in B. mori (Harshman and James, 1998; Engstrom, 1998). In Drosophila, GATA box is important for maintaining the normal expression of cecropin A1 gene in the presence of an adjacent NF-κB motif (Kadalayil et al., 1997). Drosophila Serpent, a GATA-binding transcription factor with a zinc finger domain, interacts with other proteins to induce the expression of immune proteins during embryogenesis and hematopoiesis in the adults (Tingvall et al., 2001; Waltzer et al., 2002).
In arthropods, the molting hormone 20-hydroxyecdysone (20E) regulates many biological processes including molting, embryogenesis, metamorphosis, and reproduction (Riddiford et al., 2003). Evidence indicates a linkage between insect development and immune responses. Starvation of Rodnius prolixus reduced hormone levels and compromised its immune system (Azambuja et al., 1997). Lack of ecdysteroids compromised the Drosophila cellular immune responses by negatively affecting hemocyte proliferation and encapsulation (Sorrentino et al., 2002). The promoter region of the Anopheles gambiae proPO1 gene contains two ecdysone-responsive elements (EcREs) (Ahmed et al., 1999). EcR/USP heterodimer prepared from nuclear extracts of adult A. gambiae can bind to one EcRE. Furthermore, Muller and his colleagues (1999) demonstrated 20E modulates proPO gene expressions in A. gambiae cell lines, whereas microbial and parasitic challenges did not affect A. gambiae proPO1 expression (Ahmed et al., 1999).
To understand the transcriptional regulation of the proPO activation system, we isolated the PAP-1 gene from a M. sexta genomic library and elucidated its exon–intron organization. We also identified immune- and ecdysone-responsive elements in the 5′ flanking region of the gene. In addition, we examined the PAP-1 expression patterns in different tissues and at different stages of development. The responsiveness of this immune gene to ecdysteroids was investigated as well.
2. Methods and materials
2.1. Insects
M. sexta larvae were hatched from eggs (Carolina Biological Supply) and reared as previously described (Dunn and Drake, 1983). Day 2 fifth instar larvae were injected with 2 × 108 Escherichia coli cells suspended in 100 µl phosphate saline buffer. Hemolymph and fat body samples were collected 24 h after the bacterial challenge.
2.2. Library screening, subcloning, and DNA sequencing
The 5′ PstI-SmaI fragment of PAP-1 cDNA (Jiang et al., 1998) was labelled with 32P-dCTP (3000 Ci/mmol) using Multiprime DNA Labelling System (Amersham Pharmacia Biotech). A M. sexta genomic DNA library in λGEM-11, kindly provided by Dr. Yucheng Zhu at Southern Insect Management Research Unit (USDA-ARS), was screened according to Sambrook and Russell (2001). After plaque purification and amplification, phage DNA was prepared using Wizard Lambda Preps DNA Purification System (Promega). The restriction enzyme map was determined by single and double digestions with XhoI, NcoI, XbaI, and SacI. Digested DNA fragments were separated by agarose gel electrophoresis, transferred to a nitrocellulose membrane, and hybridized with the full-length cDNA probe. Fragments of the PAP-1 gene were subcloned into pBluescript-(KS) (Stratagene). Inserts from the resulting transformants were sequenced using BigDye v2.0 Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems). Sequences were edited and analyzed using MacVector Sequence Analysis Software (Version 6.5, Oxford Molecular Ltd.).
2.3. Primer extension
Primer extension was performed by following a standard protocol (Sambrook and Russell, 1999). Primer j394 (5′-CCA ATA AAC TGC AAA CAC AAT GAA CAC-3′), corresponding to the reverse complement of nucleotides 16–42 of the PAP-1 coding region, was terminally labelled with γ-32P-ATP (3000 Ci/mmol) using T4 polynucleotide kinase. The primer (1.0 × 105 cpm) was added to 15 µg of total RNA, incubated at 60 °C for 15 min, and then slowly cooled to 30 °C for 45 min. Annealed primer-RNA complexes were extended with MMLV reverse transcriptase (200 U/µl) at 42 °C for 1 h. The extension products were analyzed on 8% polyacrylamide gels containing 7 M urea, along with DNA size standards.
2.4. Southern blot Analysis
M. sexta genomic DNA was extracted from a single fifth instar larva. Aliquots of the DNA sample (15 µg each) were incubated with restriction enzymes (SalI, SmaI, HindIII, and XbaI) at 37 °C for 6 h. The digested DNA samples were separated by electrophoresis on a 1% agarose gel and transferred to a GeneScreen Plus nitrocellulose membrane (NEN Life Science Products). The DNA blot was hybridized with 32P-labelled PAP-1 cDNA as described by Sambrook and Russell (2001).
2.5. PAP-1 mRNA level in the cultured fat body
The fat body tissue was prepared according to Kanost et al. (1995). Briefly, fat body tissues were dissected from day 2, fifth instar larvae, rinsed twice in SFM900II insect cell medium (Invitrogen Life Technologies), and transferred to separate wells of a tissue culture plate (Falcon). Each well contained 1 ml of the same medium supplemented with 10,000 U/ml penicillin G and streptomycin (Sigma). 20E (1 mg/ml in ethanol) was added to the culture to a final concentration of 1.0 or 5.0 µg/ml. The control culture was treated with the same volume of the solvent. All cultures were incubated at 27 °C with shaking at 120 rpm. Total RNA samples, extracted from the cultured fat body after 48 h, were analyzed by RT–PCR.
2.6. RNA extraction and RT–PCR analysis
Total RNA samples were prepared from different tissues of M. sexta at various developmental stages using Micro-to-Midi Total RNA Purification System (Invitrogen Life Technologies). Similarly, fat body and hemocyte RNA samples were isolated from naïve and bacteria-injected M. sexta larvae. First-strand cDNA was synthesized using total RNA (2–4 µg), oligo(dT)17 (10 pmol), and MMLV reverse transcriptase (200 U, Invitrogen Life Technologies) at 37 °C for 1 h. M. sexta ribosomal protein S3 (rpS3) transcripts were used as an internal standard to normalize the cDNA templates in a preliminary PCR experiment (Jiang et al., 2003b). Relative levels of PAP-1 cDNA in the samples (adjusted to contain an equal amount of rpS3 cDNA) were measured by semi-quantitative PCR using k628 (5′-GTC AAT ACA TAT CGC TGG TTG-3′) and j386 (5′-ATC TCC CTT TAC GAG TGC CC-3′). The thermal cycling conditions were 94 °C, 30 s; 50 °C, 30 s; 72 °C, 60 s, and the cycle numbers were chosen empirically to produce comparable band intensities while avoiding saturation. After separation on a 1.3% agarose gel electrophoresis, intensities of the PCR products were quantified and compared using Kodak Digital Science 1D Gel Analysis Software.
3. Results
3.1. Structure of M. sexta PAP-1 gene
Using the 5′ PstI-SmaI fragment of PAP-1 cDNA as a probe, we screened approximately 1 × 105 plaques and isolated one positive clone from the M. sexta genomic library. Restriction mapping and Southern blot analysis revealed that a 17.3 kb SacI-XhoI fragment included the entire PAP-1 gene (Fig. 1, upper panel). We subcloned seven restriction enzyme fragments into a plasmid vector and determined the complete nucleotide sequence of the M. sexta PAP-1 gene.
Fig. 1.
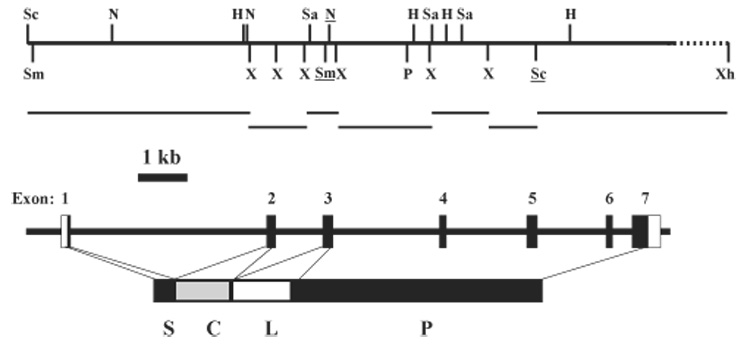
Structure of M. sexta PAP-1 gene and subcloning strategy. Upper panel: restriction map of the PAP-1 genomic insert in a positive λ bacteriophage. H, HindIII; N, NcoI; P, PstI; Sa, SalI; Sc, SacI; Sm, SmaI; X, XbaI; Xh, XhoI. The underlined recognition sites denote their presence in the cDNA. Horizontal bars mark the PAP-1 genomic fragments for subcloning. The dashed line indicates the region which is not sequenced. Lower panel: exon–intron organization of the PAP-1 gene. Numbered vertical bars: exons with the noncoding regions shown as open boxes. S, signal peptide; C, clip domain; L, linker region; P, proteinase domain. The same scale and starting site are used for the restriction map and exon–intron structure.
A comparison between the gene and cDNA sequences indicated that the PAP-1 gene is composed of seven exons and six introns (Fig. 1, lower panel). Exon 1 encodes the first 17 amino acid residues of the signal peptide. Exon 2 codes for the last two residues of the signal peptide and a complete clip domain. The subsequent 52 amino acid residues, linking the clip and proteinase domains, are encoded by 3′ end of exon 2, complete exon 3, and 5′ end of exon 4. The remaining portion of exon 4, the entire exons 5 and 6, and the 5′ coding region of exon 7 encode the SP catalytic domain. The 3′ untranslated region (226 bp) of PAP-1 cDNA is represented by the 3′ end of exon 7. We compared the 5′ and 3′ ends of the intron sequences and identified the consensus: 5′-GTRW(/A)R(/A)R(/G)TW(/A)NWV(/A) and W(/T)NW(/T)W(/A)W(/T)HHHW(/T)Y(/T)W(T)B(/T)Y(/C)Y(/C)AG-3′, where the nucleotide in each parenthesis appears in 4–5 of the six sequences at that position (Table 1). These two consensus sequences agree well with those deduced from the M. sexta serpin-1 gene (Jiang et al., 1996).
Table 1.
Intron sequences at the 5′ and 3′ splicing junctions in M. sexta PAP-1 gene
| Intron | 5′ end | 3′ end |
|---|---|---|
| 1 | GTAAGTT | TCTTTCTTTTTGCAG |
| 2 | GTAAATA | TTAAAACATTTTTAG |
| 3 | GTATGTA | TATTATTCTCATCAG |
| 4 | GTAAGTA | ACTATCATTTTCCAG |
| 5 | GTAAGTA | TTTATATTTTTTCAG |
| 6 | GTGAGTA | TGAATAACATTTCAG |
| Consensusa | GTRWRTW | WNWWWHHHWYWBYAG |
| Predominantb | GTAAGTA | T–TAT– – –TTTTCAG |
Consensus sequences were determined at each position when a particular type of nucleotide residue was present in all six positions. R: A,G; Y: C,T; W: A,T; H: A,C,T; B: C,G,T; V: A,C,G; N: A,C,G,T.
Predominant sequences were determined by the nucleotide residue present by the most frequency at the specific position.
To determine the transcription initiation site of PAP-1 gene, we carried out a primer extension experiment and detected a 167 bp product (Fig. 2). Since the primer corresponds to the reverse complement of nucleotides 16–42 of the PAP-1 coding region (Fig. 3), transcription of the gene starts at the first C of CCATG—20 nucleotides before the 5′ end of the reported cDNA (Jiang et al., 1998). Another CCAGT is present between nucleotides −7 and −3. These two sequences closely resemble the 5-nucleotide consensus sequence (TCAGT), which typically resides within 10 nucleotides before or after the transcription start site in arthropod genes (Cherbas and Cherbas, 1993). We have identified a sequence between positions −31 and −26 (TATTTA), reminiscent of the TATA or Goldberg-Hogness box (TATAAA or TATATA). There is a perfect match further upstream (TATAAA, nucleotides −115 to −110), but it is too distant from the transcription start site (Fig. 3).
Fig. 2.
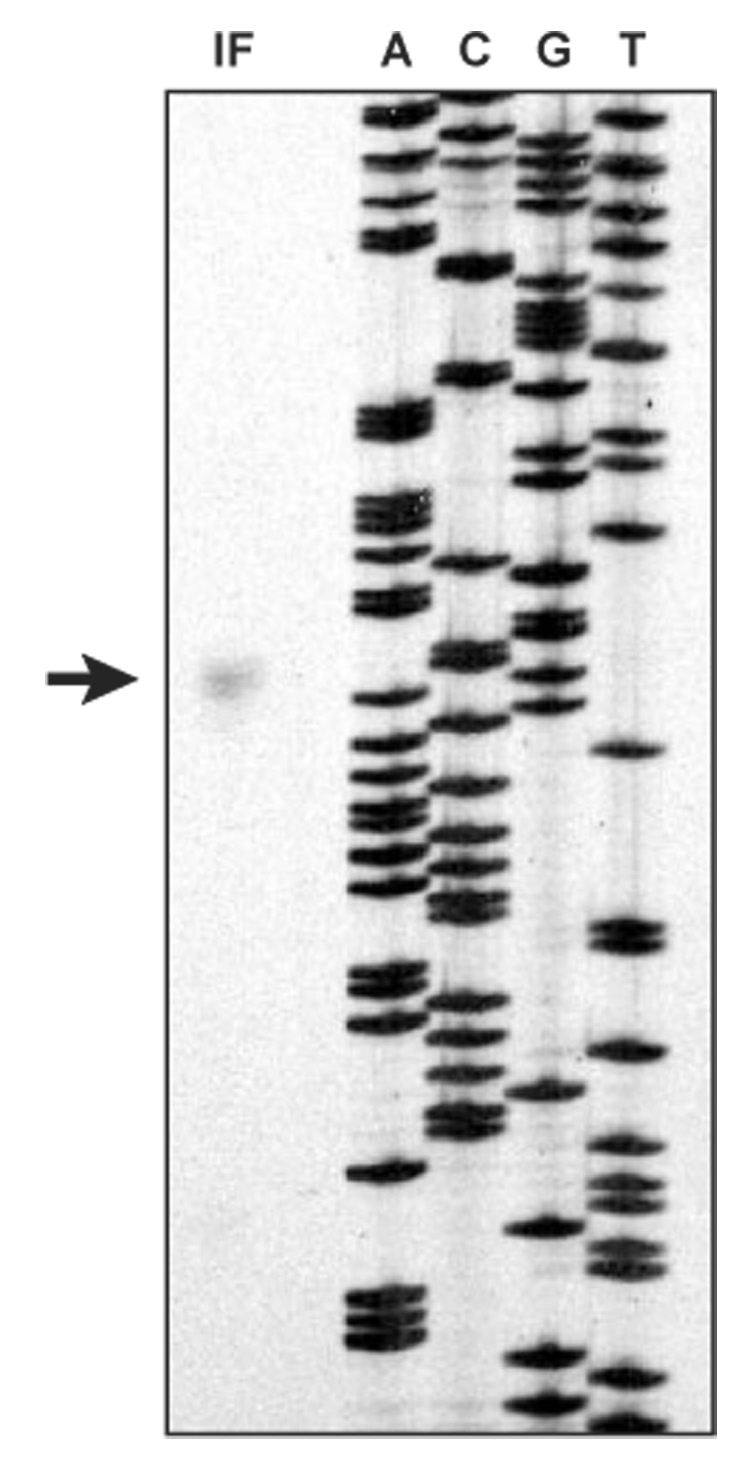
Determination of the transcription initiation site in M. sexta PAP-1 gene. A primer, complementary to nucleotides 16–42 of the PAP-1 coding region at the 3′ end of exon 1, was terminally labelled with γ-32P-dATP and annealed to total RNA from fat body from bacteria-injected larvae (15 µg). After annealing to RNA, the primer was extended with reverse transcriptase. The set of sequencing reactions (ACGT) on the right of the primer extension lane for use as a sizing ladder were from dideoxynucleotide sequencing of single-stranded M13 mp18 DNA using −40 primer. The arrow indicates the 167 bp extension product from fat body RNA isolated from the induced larvae.
Fig. 3.

Nucleotide sequence and structural features of M. sexta PAP-1 gene. Nucleotides in the 5′-flanking region are assigned negative numbers. Nucleotide 1 is assigned based on the primer extension results (Fig. 2). Exon sequences are underlined with the encoded amino acid sequences listed below translated exons, using the one-letter code under the 2nd nucleotide of each codon. While some regions of the intron sequences (marked “---”) are not shown, their sizes and positions are indicated. GATA boxes (6-nucleotide) and ISRE sites (13-nucleotide), bold and double underlined; NF-κB motifs (10-nucleotide) and EcRE (15-nucleotide), bold and single underlined; TATA boxes (6-nucleotide) and an imperfect inverted repeat (78 nucleotides), bold italic and double underlined. Mismatches in the repeat are marked with “▲”. Single nucleotide polymorphic sites are in bold italic on the DNA sequence. Among them, nonsynonymous substitutions are further indicated on the affected amino acid residues (bold and underlined). The Cys residues in the clip domain and the catalytic residues in the SP domain are indicated with “+” and “#”, respectively.
The exons are 98% identical in sequence to the PAP-1 cDNA isolated from a M. sexta larval hemocyte library (Jiang et al., 1998). Within the open reading frame, we have identified 20 nucleotide differences, most of which are synonymous substitutions. Two amino acid changes (R² to K², I5 to T5) occur in the signal peptide, and three others (A28 to V28, A286 to V286, H343 to N343) are present in the mature protein.
To examine whether there is more than one copy of the M. sexta PAP-1 gene, we isolated the genomic DNA and carried out a Southern blot analysis (Fig. 4). The number, size and intensity of the hybridizing restriction fragments were consistent with those predicted from the restriction map (Fig. 1). The SalI-digested genomic DNA generated at least four fragments at 0.6, 0.9, 2.4, and 7 kb. HindIII digestion gave rise to five hybridized fragments at about 0.7, 2.5, 3.4, 8 and > 12 kb. SmaI cut the PAP-1 gene once but did not result in hybridizing bands smaller than 6 kb, whereas XbaI yielded five hybridizing bands at 0.5, 0.6, 1.2, 3.4 and > 12 kb positions. No additional bands were observed when we used low-stringency conditions for hybridization and washing. These results indicate that the M. sexta genome may contain a single copy of the PAP-1 gene and that the nucleotide differences in the genomic sequences (Fig. 3) are most likely caused by allelic variations.
Fig. 4.
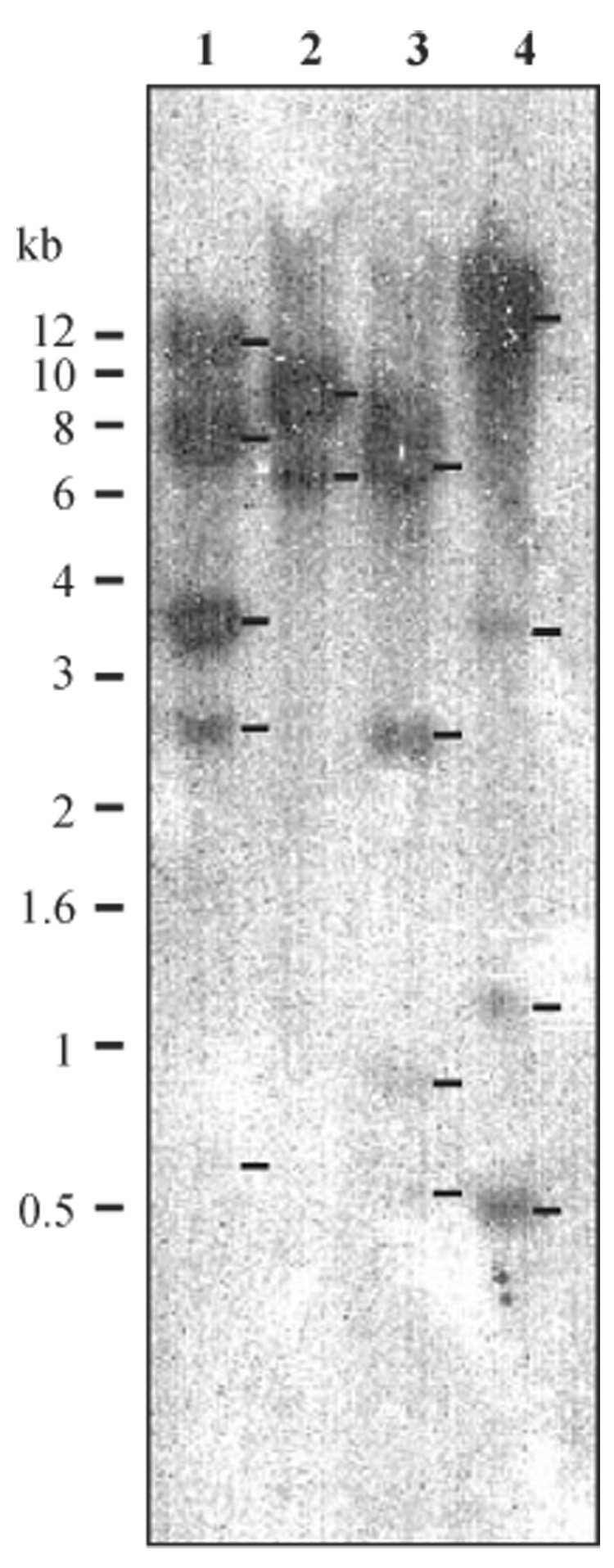
Southern blot analysis of M. sexta genomic DNA using 32P-labelled PAP-1 cDNA. Samples of the genomic DNA (15 µg) were digested with HindIII (lane 1), SmaI (lane 2), SalI (lane 3), or XbaI (lane 4). After separation by agarose gel electrophoresis and transfer to a nitrocellulose membrane, the DNA fragments were hybridized with the probe. The positions and sizes of the molecular markers are indicated on the left side.
3.2. Putative regulatory elements in the PAP-1 gene
We discovered an inverted repeat in the first intron (nucleotides 3327–3404) (Fig. 3). This region, with three mismatches in the middle, may form a 78 bp stem-loop in the PAP-1 pre-mRNA or a cruciform in the double-stranded DNA of its gene. Conceivably, such a structure may influence RNA splicing of intron 1 (4.0 kb). The lack of mismatches in this long inverted repeat suggests that there is some selective pressure to preserve its unknown structure and function.
Computer analysis of the 5′ flanking sequence allows us to locate potential regulatory elements in the PAP-1 gene (Table 2). Three NF-κB motifs are present at positions −432, −138, and −76, two of which are located on the plus strand. In comparison to the insect NF-κB consensus (Hultmark, 1993), there are one or two mismatches in these 10-nucleotide-long sequences. We have also identified three perfect GATA boxes at −618, −266, and −138. Like the NF-κB motifs, these conserved sequences frequently exist in the promoter regions of insect immune genes. There are two interferon-stimulated response elements (ISREs) at positions −58 and −120. These two motifs, both located on the plus strand, contain one or two nucleotides that mismatch the 13-base mammalian ISRE consensus. Moreover, we have identified a putative EcRE at + 1098 in the first intron. Like the 15-nucleotide consensus, this sequence (GGGTA̳CAGTGT̳ACCC) is a perfect inverted repeat—the two mismatches (double underlined) can still form a base pair.
Table 2.
Sequence analysis of the 5′ flanking region of M. sexta PAP-1 gene
| Motif name and consensusa | Sequence foundb | Location | Matched |
|---|---|---|---|
| NF-κB: GGGRAYYYYY | aGGtATTCTT(+) | −432 | 8/10 |
| aGGAATTTTT(−) | −138 | 9/10 | |
| GcGAACgCCT(+) | −76 | 8/10 | |
| GATA: WGATAA | TGATAA(+) | −618 | 6/6 |
| AGATAA(+) | −266 | 6/6 | |
| TGATAA(+) | −138 | 6/6 | |
| ISRE: GGAAANNGAAANN | GGAAATAtAAAAA(+) | −120 | 12/13 |
| GGAAcAAcAAAAC(+) | −58 | 11/13 | |
| EcRE: RRGKTCANTGAMCYY | GGGTaCAGTGtACCC(+) | 1098 | 13/15 |
R: A,G; Y: C,T; W: A,T; H: A,C,T; K: G,T; M: A,C. S: C,G; W: A,T; N: A,C,G,T.
Nucleotides in the lower case do not match with the consensus.
Assuming that immune-responsive genes are regulated similarly in M. sexta, we compared the 5′ flanking regions of PAP-1, lysozyme and hemolin genes to identify species-specific elements. The PAP-1 and hemolin sequences are 33% identical whereas the identity between PAP-1 and lysozyme sequences is 47%. We have identified 24 NF-κB, ISRE, and GATA elements in these sequences, nine of which have 0–1 mismatch as compared with the consensus motifs (Fig. 5A). However, we did not detect any regulatory elements in the corresponding regions of these sequences. The only exception was the TATA box, which is located at −29 to −31 of the three M. sexta immune genes.
Fig. 5.

Multiple sequence comparison. Panel A: 5′-flanking regions of M. sexta immune genes. The promoter regions of M. sexta lysozyme and hemolin genes (Mulnix and Dunn, 1994; Wang et al., 1995) were retrieved from Genbank and compared with the same region in the PAP-1 gene using ClustalW program (Thompson et al., 1994). Positions identical in all four sequences are marked with “*”. GATA boxes (6-nucleotide, marked G) and ISRE sites (13-nucleotide, marked I), bold and double underlined; NF-κB motifs (10-nucleotide, marked N), bold and single underlined; TATA boxes (6-nucleotide), bold italic and double underlined. Mismatches are in lower case and the motifs with 0–1 mismatch are indicated. Panel B: a highly similar intron sequence in M. sexta PAP-1, juvenile hormone-binding protein (JHBP), and eclosion hormone (EH) genes. Based on a BLASTN search, similar sequences in M. sexta PAP-1 (nucleotides 6621–6777), JHBP (nucleotides 6096–6252, AF527636), and EH (nucleotides 3468–3322, M27808) genes were retrieved from Genbank and aligned. Positions identical in all three sequences are marked with “*”.
When Genbank was searched with the PAP-1 intron sequences, we found that two M. sexta sequences contain a region closely similar to a 156-bp region at the end of intron 3 (Fig. 5B). These include juvenile hormone-binding protein and eclosion hormone genes. The sequence identities among these regions range from 80% to 86%. It is unclear whether these sequences have a regulatory function or not.
3.3. Transcription of the PAP-1 gene
We employed RT–PCR to examine the PAP-1 expression profile in different tissues at development stages. In the control reactions, we amplified an expected 0.42 kb PCR product from normalized total RNA samples (Fig. 6C). Relative band intensities indicated that PAP-1 mRNA levels in fat body and hemocytes of bacteria-injected larvae were significantly higher than those of the naïve insects. This result, consistent with our previous data from Northern blot analysis (Jiang et al., 1998) and prediction of the immune-responsive elements, validated the RT–PCR method for estimating the transcript levels.
Fig. 6.
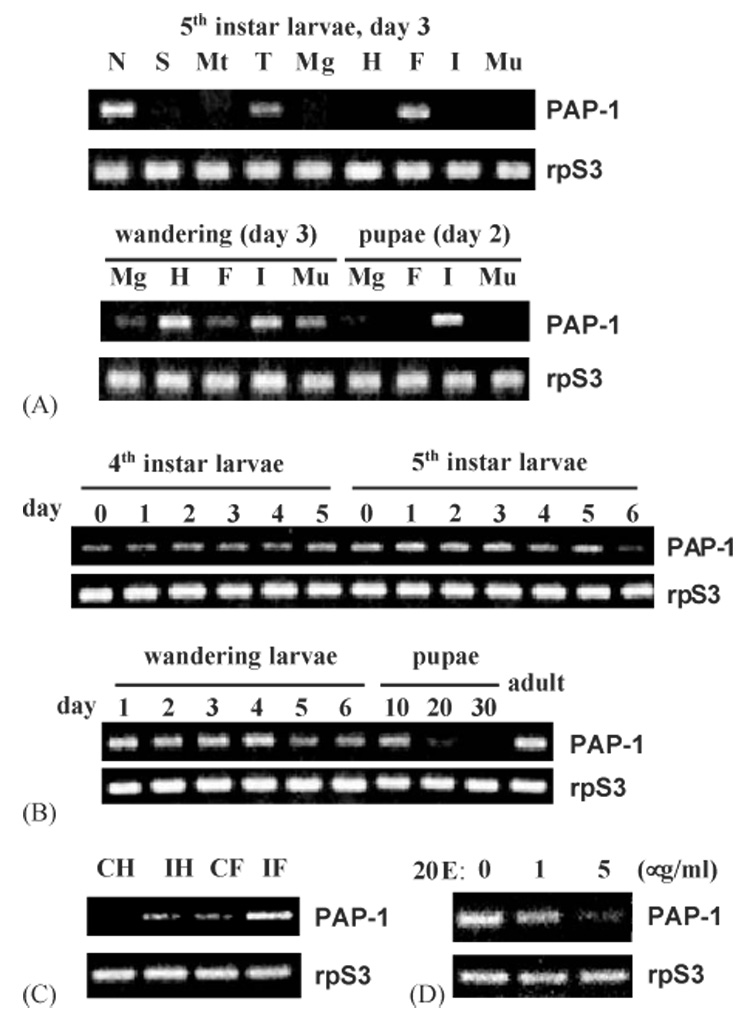
RT–PCR analysis of M. sexta PAP-1 mRNA levels. Panel A: PAP expression in nerve tissues (N), salivary glands (S), Malpighian tubules (Mt), trachea (T), midgut (Mg), hemocytes (H), fat body (F), integument (I), and muscle (Mu) of the 5th instar larvae (day 3), wandering larvae (day 3) or early pupae (day 2). Panel B: changes of PAP-1 mRNA level in fat body from M. sexta at different developmental stages. Panel C: induced transcription of PAP-1 gene in hemocytes and fat body upon bacterial infection. CH and CF: hemocytes and fat body from the naïve larvae; IH and IF: hemocytes and fat body collected from the larvae at 24 h after injection of E. coli ; Panel D: transcriptional suppression of PAP-1 gene in the cultured fat body by 20-hydroxyl ecdysone at different concentrations. M. sexta ribosomal protein S3 (rpS3) transcripts were normalized in all these analyses.
PAP-1 mRNA was detected in trachea, fat body, and the nerve cord from day 3, 5th instar naïve larvae (Fig. 6A). It was also abundantly present in integument and hemocytes from day 3, wandering larvae. Lower levels of the transcripts were identified in the other tissues at the same developmental time. In the early pupae (day 2), we detected the PAP-1 mRNA in integument, as well as midgut and fat body. Since the PAP-1 mRNA was detected in fat body samples at all these stages, we decided to closely monitor its level in fat body of M. sexta at different developmental stages.
We found that PAP-1 gene was transcribed in fat body throughout the 4th, 5th, and wandering stages (Fig. 6B). The fat body mRNA levels were higher in days 1–3 of the 5th instar and days 1–4 of the wandering larvae. In the fat body samples collected from the pupae, PAP-1 mRNA significantly decreased with time — it totally disappeared in the late pupae (day 30). The transcripts resurged in the adult fat body tissue.
The identification of a putative EcRE in intron 1 led us to test whether transcription of this immune gene is also regulated by ecdysteroids. We treated the cultured larval fat body with 20E at 0, 1, and 5 µg/ml and detected a concentration-dependent reduction in PAP-1 transcript level (Fig. 6D). While there was a significant decrease in the mRNA level at 1 µg/ml 20E, the transcripts almost completely disappeared at 5 µg/ml.
4. Discussion
So far, our knowledge of the transcriptional regulation of the insect proPO activation system is in a large part limited to proPO genes. Dimopoulos et al. (1997) found that the mRNA levels of six A. gambiae proPO did not significantly change after a bacterial challenge, while the production of GNBP and defensin was upregulated. An infection of the microfilaria worm D. immitis failed to induce proPO gene expression in the mosquito A. subalbatus (Cho et al., 1998). Neither was a proPO level change observed in A. gambiae 4a-3B cells after an inoculation of W. bancrofti microfilaria (Dimarcq et al., 1997). This unresponsiveness to pathogen or parasite infection suggests that the regulation of proPO transcription is different from those of other acute-phase genes (Dimopoulos et al., 1997, 2001). We propose that several factors may contribute to such an anomaly: (1) proPO concentration in insect hemolymph is sufficiently high, so further increase is unnecessary for an immune response against secondary infection; (2) proPO activation is regulated as a local reaction, so only a small amount of active PO is generated near the site of invasion; (3) Other PO-mediated physiological processes (i.e. molting) play more important roles in the control of the proPO expression.
The regulation of M. sexta PAP-1 transcription is different from that of the proPO expression. PAP-1 is an immune-responsive gene, whose transcripts and translation products increase significantly after a bacterial challenge (Fig. 5C and Jiang et al., 1998). Unlike M. sexta proPO, whose synthesis only occurs in oenocytoids (Jiang et al., 1997), PAP-1 transcripts were detected in several tissues/cells at different developmental stages (Fig. 5A and B). Perhaps there is a more complex mechanism for regulating the PAP-1 expression.
PAP-1 and the proPO expression also appear to be related — ecdysteroids affect the transcription of both genes. In an A. gambiae cell-line, 20E treatment up-regulates the gene transcription of proPO1–proPO4 and proPO6 (Ahmed et al., 1999; Muller et al., 1999). In contrast, the proPO5 expression was suppressed in the cells after an exposure to 20E. RT–PCR analysis of their transcripts in the mosquito yielded a similar result: proPO1–proPO4 and proPO6 mRNA became more abundant after a blood meal whereas proPO5 mRNA decreased. Analogous to the mosquito proPOs, multiplePAPs are present in the tobacco hornworm. M. sexta PAP-1 gene is likewise under the control of 20E. It is known that 20E and juvenile hormone regulate the M. sexta metamorphosis and ovary maturation (Riddiford et al., 2003). There are two ecdysteroid peaks in the plasma of the late 5th instar and late wandering larvae. Consistent with the result from RT–PCR analysis of the cultured fat body (Fig. 5D), PAP-1 mRNA levels in these two periods were much lower than those in the early 5th instar and early wandering larvae (when the 20E titers were low) (Fig. 5B). The major ecdysone peak in the middle pupal stage also coincided with the drastic reduction of PAP-1 mRNA level in the fat body. Certainly, further experiments are needed to elucidate physiological role of ecdysone-mediated suppression of PAP-1 expression and to validate if the putative EcRE in intron 1 is indeed involved in the down-regulation. Furthermore, it would be interesting to examine if the highly similar intron sequences in the PAP-1, juvenile hormone-binding protein, and eclosion hormone genes (Fig. 5B) play a role in their transcriptional regulation. In addition, immune and hormonal responsiveness and transcriptional profiles of M. sexta PAP-2 and PAP-3 genes are worth investigating.
To identify DNA sequences that might account for its immune responsiveness, we searched the 5′ flanking sequence of the M. sexta PAP-1 gene. We found three sequences with similarity to the NF-κB consensus (Table 2 and Fig. 3). Three GATA boxes, two ISREs, and one 78-bp inverted repeat were identified. Further analyses are needed to test whether any of these putative elements are involved in the up-regulation of PAP-1 expression upon microbial infection.
PAP-1 gene, spanning nearly 13,000 nucleotides, consists of seven exons. Compared to the clip-domain SPs from D. melanogaster and A. gambiae, the exon number and gene length are significantly larger (data not shown). The clip domain in PAP-1 is entirely encoded by exon 2. In the putative Drosophila PAP genes (De Gregorio et al., 2001), the clip domain is also encoded by a single exon in SP4, SP7 and SP10 genes (Ross et al., 2003). These results suggest that the evolution of clip domains could result from exon shuffling. In contrast, the clip domain in Drosophila SP25 (another predicted PAP) is encoded by two exons. The clip domains in Drosophila SPs and SPHs are dissimilar in their sequences, except for the conserved Cys residues. These observations indicate that the evolutionary history of clip domains is complex and has to be examined individually.
Acknowledgments
This work was supported by the National Institutes of Health Grant GM58634 (to H.J). We thank Drs. Melcher and Dillwith for their helpful comments on the manuscript. This article was approved for publication by the Director of Oklahoma Agricultural Experimental Station and supported in part under project OKLO2450. The nucleotide sequence reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number AY789465.
Abbreviations
- PO and proPO
phenoloxidase and its precursor
- PAP
proPO-activating proteinase
- RT–PCR
reverse transcription–polymerase chain reaction
- 20E
20-hydroxyecdysone
- EcRE
ecdysone-responsive element
- NF
nuclear factor
- ISRE
interferon-stimulated response element
- SP and SPH
serine proteinase and its homolog
References
- Ahmed A, Martin D, Manetti AG, Han SJ, Lee WJ, Mathiopoulos KD, Muller HM, Kafatos FC, Raikhel A, Brey PT. Genomic structure and ecdysone regulation of the prophenoloxidase 1 gene in the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. USA. 1999;96:14795–14800. doi: 10.1073/pnas.96.26.14795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida M, Brey PT. Recent advances on the research of the insect prophenoloxidase cascade. In: Brey PT, Hultmark D, editors. Molecular Mechanisms of Immune Responses in Insects. London: Chapman & Hall; 1998. pp. 135–172. [Google Scholar]
- Azambuja P, Garcia ES, Mello CB, Feder D. Immune responses in Rhodnius prolixus: influence of nutrition and ecdysone. J. Insect Physiol. 1997;43:513–519. doi: 10.1016/s0022-1910(97)00010-3. [DOI] [PubMed] [Google Scholar]
- Cherbas L, Cherbas P. The arthropod initiator: the capsite consensus plays an important role in transcription. Insect Biochem. Mol. Biol. 1993;23:81–90. doi: 10.1016/0965-1748(93)90085-7. [DOI] [PubMed] [Google Scholar]
- Cho WL, Liu HS, Lee CH, Kuo CC, Chang TY, Liu CT, Chen CC. Molecular cloning, characterization and tissue expression of prophenoloxidase cDNA from the mosquito Armigeres subalbatus inoculated with Dirofilaria immitis micro-filariae. Insect Mol. Biol. 1998;7:31–40. doi: 10.1046/j.1365-2583.1998.71049.x. [DOI] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. USA. 2001;98:12590–12605. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimarcq JL, Imler JL, Lanot R, Ezekowitz RA, Hoffmann JA, Janeway CA, Lagueux M. Treatment of 1(2)mbn Drosophila tumorous blood cells with the steroid hormone ecdysone amplifies the inducibility of antimicrobial peptide gene expression. Insect Biochem. Mol. Biol. 1997;27:877–886. doi: 10.1016/s0965-1748(97)00072-6. [DOI] [PubMed] [Google Scholar]
- Dimopoulos G, Richman A, Muller HM, Kafatos FC. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc. Natl. Acad. Sci. USA. 1997;94:11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos G, Muller HM, Levashina EA, Kafatos FC. Innate immune defense against malaria infection in the mosquito. Curr. Opin. Immunol. 2001;13:79–88. doi: 10.1016/s0952-7915(00)00186-2. [DOI] [PubMed] [Google Scholar]
- Dunn P, Drake D. Fate of bacteria injected into naïve and immunized larvae of the tobacco hornworm, Manduca sexta. J. Invertebr. Pathol. 1983;41:77–85. [Google Scholar]
- Engstrom Y. Insect immune gene regulation. In: Brey PT, Hultmark D, editors. Molecular Mechanisms of Immune Responses in Insects. London: Chapman & Hall; 1998. pp. 211–244. [Google Scholar]
- Harshman LG, James AA. Differential gene expression in insects: transcriptional control. Ann. Rev. Entomol. 1998;43:671–700. doi: 10.1146/annurev.ento.43.1.671. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- Huang TS, Wang H, Lee SY, Johansson MW, Söderhäll K, Cerenius L. A cell adhesion protein from the crayfish Pacifastacus leniusculus, a serine proteinase homologue similar to Drosophila masquerade. J. Biol. Chem. 2000;275:9996–10001. doi: 10.1074/jbc.275.14.9996. [DOI] [PubMed] [Google Scholar]
- Hultmark D. Immune reactions in Drosophila and other insects: a model for innate immunity. Trends Genet. 1993;9:178–183. doi: 10.1016/0168-9525(93)90165-e. [DOI] [PubMed] [Google Scholar]
- Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochem. Mol. Biol. 2000;30:95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Huang Y, Mulnix AB, Kadel J, Cole K, Kanost MR. Organization of serpin gene-1 from Manduca sexta: evolution of a family of alternate exons encoding the reactive site loop. J. Biol. Chem. 1996;271:28017–28023. doi: 10.1074/jbc.271.45.28017. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Ma C, Kanost MR. Subunit composition of pro-phenol oxidase from Manduca sexta: molecular cloning of subunit proPO-p1. Insect Biochem. Mol. Biol. 1997;27:835–850. doi: 10.1016/s0965-1748(97)00066-0. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Kanost MR. Pro-phenoloxidase activating proteinase from an insect, Manduca sexta: a bacteria-inducible protein similar to Drosophila easter. Proc. Natl. Acad. Sci. USA. 1998;95:12220–12225. doi: 10.1073/pnas.95.21.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Yu XQ, Kanost MR. Prophenoloxidase-activating proteinase-2 from hemolymph of Manduca sexta: a bacteria-inducible serine proteinase containing two clip domains. J. Biol. Chem. 2003a;278:3552–3561. doi: 10.1074/jbc.M205743200. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Yu XQ, Zhu Y, Kanost M. Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem. Mol. Biol. 2003b;33:1049–1060. doi: 10.1016/s0965-1748(03)00123-1. [DOI] [PubMed] [Google Scholar]
- Kadalayil L, Petersen UM, Engstrom Y. Adjacent GATA and kappa B-like motifs regulate the expression of a Drosophila immune gene. Nucleic Acids Res. 1997;25:1233–1239. doi: 10.1093/nar/25.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanost MR. Serine proteinase inhibitors in arthropod immunity. Dev. Comp. Immunol. 1999;23:291–301. doi: 10.1016/s0145-305x(99)00012-9. [DOI] [PubMed] [Google Scholar]
- Kanost MR, Prasad SV, Huang Y, Willott E. Regulation of serpin gene-1 in Manduca sexta. Insect Biochem. Mol. Biol. 1995;25:285–291. doi: 10.1016/0965-1748(94)00067-r. [DOI] [PubMed] [Google Scholar]
- Lee SY, Kwon TH, Hyun JH, Choi JS, Kawabata SI, Iwanaga S, Lee BL. In vitro activation of pro-phenol-oxidase by two kinds of pro-phenol-oxidase-activating factors isolated from hemolymph of coleopteran, Holotrichia diomphalia larvae. Eur. J. Biochem. 1998;254:50–57. doi: 10.1046/j.1432-1327.1998.2540050.x. [DOI] [PubMed] [Google Scholar]
- Muller HM, Dimopoulos G, Blass C, Kafatos FC. A hemocyte-like cell line established from the malaria vector Anopheles gambiae expresses six prophenoloxidase genes. J. Biol. Chem. 1999;274:11727–11735. doi: 10.1074/jbc.274.17.11727. [DOI] [PubMed] [Google Scholar]
- Mulnix AB, Dunn PE. Structure and induction of a lysozyme gene from the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 1994;24:271–281. doi: 10.1016/0965-1748(94)90007-8. [DOI] [PubMed] [Google Scholar]
- Muta T, Hashimoto R, Miyata T, Nishimura H, Toh Y, Iwanaga S. Proclotting enzyme from horseshoe crab hemocytes: cDNA cloning, disulfide locations, and subcellular localization. J. Biol. Chem. 1990;265:22426–22433. [PubMed] [Google Scholar]
- Nappi AJ, Vass E. Cytotoxic reactions associated with insect immunity. Adv. Exp. Med. Biol. 2001;484:329–348. doi: 10.1007/978-1-4615-1291-2_33. [DOI] [PubMed] [Google Scholar]
- Riddiford LM, Hiruma K, Zhou X, Nelson CA. Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem. Mol. Biol. 2003;33:1327–1338. doi: 10.1016/j.ibmb.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Ross J, Jiang H, Kanost MR, Wang Y. Serine proteases and their homologs in the Drosophila melanogaster genome: an initial analysis of sequence conservation and phylogenetic relationship. Gene. 2003;304:117–131. doi: 10.1016/s0378-1119(02)01187-3. [DOI] [PubMed] [Google Scholar]
- Roxstrom-Lindquist K, Lindstrom-Dinnetz I, Olesen J, Engstrom Y, Faye I. An intron enhancer activates the immunoglobulin-related hemolin gene in Hyalophora cecropia. Insect Mol. Biol. 2002;11:505–515. doi: 10.1046/j.1365-2583.2002.00359.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning, A Laboratory Manual. third ed. New York: Cold Spring Harbor Laboratory; 2001. pp. 2.90–2.100. [Google Scholar]
- Satoh D, Horii A, Ochiai M, Ashida M. Prophenoloxidase-activating enzyme of the silkworm, Bombyx mori: purification, characterization, and cDNA cloning. J. Biol. Chem. 1999;274:7441–7453. doi: 10.1074/jbc.274.11.7441. [DOI] [PubMed] [Google Scholar]
- Sorrentino RP, Carton Y, Govind S. Cellular immune response to parasite infection in the Drosophila lymph gland is developmentally regulated. Dev. Biol. 2002;243:65–80. doi: 10.1006/dbio.2001.0542. [DOI] [PubMed] [Google Scholar]
- Sun SC, Faye I. Cecropia immunoresponsive factor, an insect immunoresponsive factor with DNA-binding properties similar to nuclear-factor kappa B. Eur. J. Biochem. 1992;204:885–892. doi: 10.1111/j.1432-1033.1992.tb16708.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingvall TO, Ross E, Engstrom Y. The GATA factor Serpent is required for the onset of the humoral immune response in Drosophila embryos. Proc. Natl. Acad. Sci.USA. 2001;98:3884–3888. doi: 10.1073/pnas.061230198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltzer L, Bataille L, Peyrefitte S, Haenlin M. Two isoforms of Serpent containing either one or two GATA zinc fingers have different roles in Drosophila hematopoiesis. EMBO J. 2002;21:5477–5486. doi: 10.1093/emboj/cdf545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Willott E, Kanost MR. Organization and expression of the hemolin gene, a member of the immunoglobulin superfamily in an insect, Manduca sexta. Insect Mol. Biol. 1995;4:113–123. doi: 10.1111/j.1365-2583.1995.tb00015.x. [DOI] [PubMed] [Google Scholar]
- Wang R, Lee SY, Cerenius L, Söderhäll K. Properties of the prophenoloxidase activating enzyme of the freshwater crayfish, Pacifastacus leniusculus. Eur. J. Biochem. 2001;268:895–902. doi: 10.1046/j.1432-1327.2001.01945.x. [DOI] [PubMed] [Google Scholar]


