Abstract
Microbial infection leads to proteolytic activation of Drosophila spätzle, which binds to the toll receptor and induces the synthesis of immune proteins. To test whether or not this mechanism exists in lepidopteran insects, we cloned the cDNA of Bombyx mori spätzle-1 and overexpressed the full-length and truncated BmSpz1 cDNA in Escherichia coli. The insoluble fusion proteins were affinity-purified under denaturing condition. After the silkworm larvae were injected with renatured BmSpz1, mRNA levels of antimicrobial peptide genes greatly increased. Similar transcriptional up-regulation was also found in Manduca sexta. Injection of pro-BmSpz1 had no such effect. When pro-BmSpz1 and Micrococcus luteus were incubated with the plasma from M. sexta larvae, we detected proteolytic processing of pro-BmSpz1. These results suggest that active spätzle is required for the induced production of antimicrobial peptides in B. mori and M. sexta.
Keywords: Serine proteinase, Cytokine processing, Signal transduction, Hemolymph protein, Insect immunity, Gene regulation, Bombyx mori, M. sexta
1. Introduction
Drosophila spätzle is a key signal transducer for embryonic development and immune responses [1,2]. It is synthesized as an inactive precursor and requires a serine proteinase cascade for cleavage activation. The embryonic cascade consists of nudel, gastrulation defective, snake, and easter [3]. Spätzle, an active ligand resulted from easter-mediated proteolysis of pro-spätzle, binds to the toll receptor on the ventral side of the syncytial embryo to initiate an intracellular signaling pathway for ventralization. In Drosophila adults, pro-Spz processing and toll pathway activation induce the synthesis of immune proteins, such as antimicrobial peptides. While fungal or Gram-positive bacterial infection is known to trigger pro-spätzle activation, constituents of the proteinase cascade in the adult plasma were largely unknown until recently. Similar to the clip-domain serine proteinases snake and easter, persephone and spätzle-processing enzyme are components of the enzyme network [4-7].
While Drosophila genetic studies have provided a good model on how serine proteinase cascades mediate developmental and immune signals, it is unclear if similar systems exist in other insects. Biochemical studies in large insects, such as Bombyx mori and Manduca sexta, may provide evidence for their existence and physiological function. In 2004, the complete nucleotide sequence of the domestic silkworm was determined [8]. We searched the genome database and identified a gene for spätzle-like protein. In order to explore its role, we cloned and expressed the intact and truncated forms of B. mori spätzle-1 which mimic the inactive precursor and active ligand, respectively. The potential role of BmSpz1 in inducing antimicrobial peptide synthesis was examined. We further tested the usefullness of recombinant pro-BmSpz1 in the detection and purification of spätzle-processing proteinases from B. mori and M. sexta.
2. Materials and methods
2.1. Insect rearing, microbe injection, fat body and hemolymph collection
Silkworm eggs and artificial diet were purchased from Carolina Biological Supplies. Day 3, fifth instar silkworm larvae were injected with H2O (50 μl), Micrococcus luteus (1 mg/ml, 50 μl), formalin-killed Escherichia coli (2.0 × 109 cells/ml, 50 μl), or formalin-killed Saccharomyces cerevisiae (5.0 × 108 cells/ml, 50 μl). Fat body tissues were dissected at 24 and 40 h after injection for total RNA isolation. M. sexta eggs were ordered from the same vender and the larvae were reared as previously described [9]. Day 2, fifth instar larvae of M. sexta were injected with 50 μl H2O or 50 μl mixture of formalin-killed E. coli (8 × 108 cells/ml), M. luteus (0.2 mg/ml) and curdlan (0.2 mg/ml). Hemolymph was collected at 24 h after injection and hemocytes were removed by centrifugation.
2.2. Expression of BmSpz1 in various tissues and developmental stages
To examine the tissue specificity of BmSpz1 transcription, total RNA samples were isolated from dissected fat body, midgut, silk gland, head, integument, ovary, and testis of fifth instar silkworm larvae as previously described [10]. Eggs were dissected from oviducts of adult female for RNA extraction. Total RNA samples of the whole larvae (in the middle of 2-5th instars) and pupae (days 1, 3, 5, 8) were prepared similarly for developmental profiling. Primers F1 (5′-CGTGCCTACGTCATACAACATA-3′) and R1 (5′-AGTTTAACCGAGTAGCGTGGC-3′) were used for BmSpz1 cDNA amplification by RT-PCR. Primers specific for silkworm actin-3 (Table 1) were used in the control reactions. The cDNA fragments were amplified for 30 cycles at 94 °C for 40 s, 53 °C for 40 s, and 72 °C for 60 s, followed by a final extension at 72 °C for 10 min. The PCR products were separated by gel electrophoresis and stained with ethidium bromide.
Table 1.
Oligonucleotide primers used in RT-PCR
| Gene name | Primer | Sequence (5′-3′) | Product length (bp) |
|---|---|---|---|
| M. sexta | |||
| rpS3 | k504 | CGCGAGTTGACTTCGGT | 400 |
| k501 | GCCGTTCTTGCCCTGTT | ||
| attacin (C14) | #1 | CGTGTCGAACTTCTTAAAGCC | 288 |
| #2 | CCTCCTCCACAACAACAACC | ||
| attacin (D23) | j953 | CGACAACGTCAATGGACACG | 158 |
| j954 | GTGGAATATCCGGCATGGTC | ||
| cecropin (I46) | #A | CCGTGTTTTATTCTTCGTCTTC | 103 |
| #B | AATCCT TTGACCTGCACCC | ||
| gloverin (I7) | #F | ACTCCAAACAAGTCCGC | 425 |
| #R | TCACCATCTATGCTGGA | ||
| lebocin (F43) | j955 | CTGATTTTGGGCGTTGCGCTG | 244 |
| j956 | GCGCGTATCTTCTATCTGGA | ||
| moricin (G37) | #1 | CGCGTTGTCGCTATTATTTTC | 107 |
| #2 | TTATTGCTCGTAGACCTTTTCC | ||
| B. mori | |||
| actin-3 | j049 | GGAGCACCCCGTCCTGCTCAC | 728 |
| j050 | CGATCCATACGGAGTACTTCCT | ||
| attacin-2 | 457 | AGATGTCCAAGAGTGTAGCGT | 664 |
| 458 | CCCATTATCAAGATTATTTAGAAG | ||
| cecropin-A1 | 478 | CGGCACTATAGAATTTCGGT | 400 |
| 479 | GGCAATGACTGTGGTATTCTTAT | ||
| cerropin-B1 | 481 | GCGCCGCTTGTGTCTTAAC | 290 |
| 482 | GATTAAATAATACTAGTATTTATGGCAGT | ||
| Cercropin-D1 | 488 | GAATTCGAAAAATGAAAATCTCG | 210 |
| 489 | ATAATTTAAGCTCTATCCTTGTCCG | ||
| gloverin-B | 449 | CCTCGCGATATTCACGACTT | 337 |
| 450 | CGGCTGACAAGTGAGTGTTC | ||
| gloverin-A5 | 455 | CAACTCAAAATGAATTCCAAATTGC | 484 |
| 456 | TGTGACCAAATTCCTTCGAGACC | ||
| lebocin-3 | 463 | GTTCTTTGCTCAGGCTTCG | 421 |
| 464 | TCGGTAACGGTTTCCCAT | ||
| moricin-A1 | 492 | TGGCAATGTCTCTGGTGT | 291 |
| 493 | GTAAGTACTACACAGGGT |
2.3. Cloning of BmSpz1 cDNA and construction of expression plasmids
Based on the initial gene prediction, Primers F2 (5′-TCGCCTCACAGTCACCCA-3′) and R2 (5′-GATCATTTCGCCGCTTTC-3′) were synthesized for PCR amplification of a BmSpz1 cDNA fragment using fat body mRNA from B. mori larvae injected with lipopolysaccharide (10 μl, 1 mg/ml). The thermal cycling conditions were 35 cycles of 94 °C, 30 s; 57 °C, 30 s; and 72 °C, 45 s, followed by 10 min incubation at 72 °C. The reaction product was cloned into pMD18-T vector (TakaRa) and confirmed by DNA sequence analysis.
To express pro-BmSpz1 in E. coli, a second PCR was performed using BmSpz1/pMD18 and primers j244 (5′-GACCCATGGACCAGCAGGATTCGCCTCACAGTCACC-3′) and j245 (5′-CAAGAGCTCGGAACTTTCATGGACCTCTCGATCATTTCGCCGCT-3′), which extended the partial cDNA in both ends. The thermal cycling conditions were 30 cycles of 94 °C, 20 s; 50 °C, 40 s; and 68 °C, 60 s, followed by 10 min incubation at 68 °C. After sequence verification, the PCR product was digested with NcoI and SacI, separated by gel electrophoresis, and recovered using QIAquick Gel Extraction Kit (Qiagen). To put in the missing part at the 3′ end, two oligonucleotides, j246 (5′-GTTGTTCGTGTGTTGCCACGCTACTCGGTTA-3′, 0.3 nmol) and j247 (5′-AGCTTAACCGAGTAGCGTGGCAACACACGAACAACAGCT-3′, 0.3 nmol) were treated with T4 polynucleotide kinase (2.0 U, Promega), 1 mM ATP and buffer in a total volume of 50 μl at 37 °C for 1 h. After denaturation for 5 min at 95 °C, the reaction mixture was slowly cooled down to 30 °C in 2-3 h. The annealed fragment, containing the adhesive ends of SacI and HindIII, was ligated with the NcoI-SacI fragment of BmSpz1 cDNA and Tev-H6pQE60 [11] cleaved by NcoI and HindIII. Insert size, restriction pattern, and DNA sequence were examined using plasmids isolated from transformants that produced 31 kDa pro-BmSpz1.
In order to express the truncated spätzle in E. coli, the cDNA in pro-BmSpz1 /Tev-H6pQE60 was used as a template to amplify a 360 bp fragment using Advantage DNA polymerase mix (Clontech), primer j253 (5′-CCCATGGCAGGCTCATTCGAAGACTC-3′) and vector-specific primer j028 (5′-GATCTATCAACAGGAGTCCA-3′). Primer j253 corresponds to nucleotides 680-705 of the cDNA, except for a few modifications to introduce an NcoI site. The thermal cycling conditions were 35 cycles of 95 °C, 30 s; 50 °C, 30 s; and 68 °C, 60 s. The PCR product was digested with NcoI-HindIII and directionally cloned into the same sites of H6pQE60 [11].
2.4. Expression, purification and renaturation of recombinant pro-BmSpz1 and BmSpz1
Fresh single colonies of E. coli JM109 carrying pro-BmSpz1/TevH6pQE60 or BmSpz1 /H6pQE60 were inoculated in 2 × YT medium supplemented with 0.1% glucose and 100 μg/ml ampicillin (pH 7.6, 3 ml/tube, 10 tubes) at 37 °C for 12 h with shaking at 300 rpm. The cultures were transferred to 1.01 of the same medium (500 ml in 2.81 flask, 2 flasks) and grown for about 3.5 h until A600 reached 0.8. Isopropyl-β-d-thiogalactopyranoside was then added to a final concentration of 1.0 mM, and the cultures continued to grow at 37 °C for 4.5 h with shaking.
The E. coli cells were harvested by centrifugation at 4500g for 20 min, resuspended in 30 ml buffer A (50 mM sodium phosphate, 0.3 M NaCl, 0.02% Tween-20, pH 8.0) containing 1.0 mg/ml lysozyme, and stirred on ice for 1 h. Following sonication (60 s/burst at 350 W for 10 times), the debris was separated from soluble fraction by centrifugation at 40,000g for 30 min. After thorough washing with buffer A supplemented with 1 M urea, the cell pellet containing insoluble pro-BmSpz1 or BmSpz1 was dissolved in 15 ml buffer B (8 M urea, 0.1 M NaH2PO4, 10 mM Tris, pH 8.0). Undissolved components were removed by ultracentrifugation at 245,000g for 30 min. The supernatant was collected and incubated with 2.0 ml Ni-NTA agarose slurry (Qiagen) under gentle agitation for 1 h at 4 °C. The suspension was then loaded into an empty poly-prep column (Bio-Rad) and washed with 10 ml buffer B, 10 ml buffer C (8 M urea, 0.1 M NaH2PO4, 10 mM Tris, pH 6.3), and 10 ml buffer D (8 M urea, 0.1 M NaH2PO4, 10 mM Tris, pH 5.9). The recombinant protein was then eluted with 10 ml buffer E (8 M urea, 0.1 M NaH2PO4, 10 mM Tris, pH 4.5). One milliliter fractions were collected and analyzed by SDS-PAGE.
The purified proteins were renatured by dialysis against 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2 mM reduced glutathione, 0.2 mM oxidized glutathione, and 5% glycerol with decreasing concentrations of urea (4.0, 2.0, and 1.0 M). The final buffer was 20 mM Tris-HCl, pH 7.5, and 100 mM NaCl. Each dialysis step was performed at 4 °C for 12 h.
2.5. Antiserum preparation and mass determination
Pro-BmSpz1 (500 μg) in the renaturation buffer with 2.0 M urea was concentrated on a Centriprep-10 (Amicon) for use as an antigen in the production of a polyclonal rabbit antiserum (Cocalico Biologicals Inc.) The molecular masses of pro-BmSpz1 and BmSpz1 were measured by MALDI-TOF mass spectrometry [12]. For mass fingerprint analysis, pro-BmSpz1 (1.0 μg) was separated by SDS-PAGE under the reducing condition and stained with Coomassie blue. The protein bands were subjected to in-gel trypsin digestion and mass determination [13].
2.6. Protein injection, RNA isolation, and RT-PCR analysis
Day 3, fifth instar B. mori larvae and day 2, fifth instar M. sexta larvae were injected with water (50 μl), BmSpz1 (0.1 mg/ml, 50 μl) or pro-BmSpz1 (0.2 mg/ml, 50 μl). Fat body tissues were dissected from the larvae at 24 h (M. sexta) and 40 h (B. mori) after injection for total RNA extraction using Micro-to-midi total RNA purification system (invitrogen). In the RT-PCR experiments, RNA samples (4 μg), oligo dT (0.5 μg) and dNTPs (10 mM each, 1 μl) were mixed with RNase-free H2O in a final volume of 12 μl, denatured at 65 °C for 5 min, and quickly chilled on ice for 5 min. cDNA was synthesized at 37 °C for 50 min by MMLV reverse transcriptase (1 μl) in the presence of dithiothreitol (0.1 M, 2 μl), RNase OUT (40 U/μl, 1 μl, Ambion), 5 × buffer (4 μl), and the denatured RNA sample (12 μl). B. mori actin-3 and M. sexta rpS3 transcripts were used as internal standards to normalize the cDNA pools in the preliminary PCR experiments. Antimicrobial peptide cDNA fragments were amplified using primer pairs specific for the genes (Table 1). The thermal cycling conditions were 94 °C, 30 s; 53 °C, 30 s; 72 °C, 50 s, and the cycle numbers were chosen empirically to produce comparable band intensities while avoiding saturation. After electrophoretic separation on a 1.5% agarose gel, intensities of bands at the correct sizes were quantified and compared using Kodak Digital Science Gel Analysis Software.
2.7. Cleavage of pro-BmSpz1 by proteinases in M. sexta hemolymph
The silkworm spätzle precursor (0.2 μg, 1 μl), plasma (2 μl) from bacteria-injected M. sexta or B. mori larvae, M. luteus (1 μl, 1 mg/ml) and buffer (20 mM Tris, pH 8.0, 10 μl) were incubated at room temperature for 15 min. The reaction mixture (11 μl) was analyzed by 15% SDS-PAGE followed by immunoblotting to reveal possible proteolytic cleavage of pro-BmSpz1.
3. Results
3.1. cDNA sequence, gene structure, and protein features
Our search of the silkworm genome database (http://silkworm.genomics.org.cn/) using Drosophila spätzle-1 indicated that Scaffold 002682 contains gene segments for a spätzle-like protein. These segments correspond to the last four exons of B. mori spätzle-1 gene. Genbank search with the coding sequence resulted in four ESTs, later assembled into the full-length BmSpz1 cDNA (Fig. 1A). The 1289-nucleotide sequence includes an 834 bp open reading frame coding for a 277-residue polypeptide. Comparison of the cDNA and scaffold revealed the exon-intron organization of BmSpz1, as well as problems with the assembled genomic sequence. The silkworm spätzle-1 gene consists of six exons and five introns. There are two copies of exon 2 located at nucleotides 13,905-14,140 and 24,054-24,289—the 827 bp flanking intron sequences are also identical. Exons 3-6 are located on the reverse complement strand of nucleotides 26,704-36,536.
Fig. 1.
cDNA sequence and gene structure of BmSpz1. (A) Nucleotide and deduced amino acid sequences of B. mori spätzle-1. Amino acid residues, shown in one-letter abbreviations, are aligned with the second nucleotide of each codon. The signal peptide and primer-binding regions are underlined. The exon boarders are indicated by “|”. Putative N- and O-linked glycosylation sites are marked with “◆” and “■”, respectively. The nucleotide differences (shaded) between cDNA and gene lead to nonsynonymous substitutions (I87 to K87 and T171 to A171). The predicted cleavage (*) occurs between R170 (bolded and underlined) and A171. (B) Putative regulatory elements in the upstream region of BmSpz1. GATA boxes (double underlined), NF-κB motifs (boldface italic), and interferon-stimulated response elements (underlined) with mismatches shown in lower case. Exon 1, starting in the middle of GCAGT, is underlined. (C) Sequence comparison of pro-BmSpz1 (Bm) and Drosophila pro-spätzle (Dm). Following the signal peptide (underlined), the mature proteins contain nine Cys residues (bold). The paired numbers (1-1, 2-2, 3-3, 4-4) indicate the predicted intrachain disulfide linkages, whereas Cys5 may form an intermolecular disulfide bond with its counterpart in another subunit. *, identical; “:” and “.”, similar. The putative proteolytic activation site is right after R219 (in DmSpz1) and R170 (in BmSpz1) (bolded and underlined).
The cDNA 5′ end (AGT) may correspond to the transcription initiation site of BmSpz1 because this AGT is in the context of GCAGT (Fig. 1B), a sequence closely similar to the five-nucleotide consensus sequence (TCAGT) typically located within 10 nucleotides before or after the start site in arthropod genes [14]. There is no TATA box (TATAAA or TATATA) near position -30. Computer analysis of the 1.5 kb 5′ flanking sequence has revealed 14 NF-κB motifs, four interferon-stimulated response elements and three perfect GATA boxes. In comparison to the insect NF-κB consensus [15], there are two mismatches in these sequences. Exon 1 consists of a ≥ 175 bp 5′ untranslated region and a 39 bp sequence coding for residues 1-13 of the signal peptide. Exon 2 (235 bp) encodes the last five residues of the signal peptide and the first half of the pro-region. Exon 3 (166 bp) and part of exon 4 (218 bp) encode the rest of the pro-region ending with Ile-Ala-Gln-Arg170. Thus, a clip-domain serine proteinase with trypsin-like specificity may activate pro-BmSpz1 by limited proteolysis right after Arg170. The active ligand, starting with Ala171-Gly-Ser-Phe, is coded by 3′ end of exon 4, entire exon 5 (101 bp), and 5′ end of exon 6 (328 bp). The 3′ end of the last exon corresponds to the 3′ untranslated region in the cDNA.
Sequence alignment indicates BmSpz1 is 54% similar in amino acid sequence to Drosophila spätzle-1 (Fig. 1C). These two sequences and spätzle-1 s of Aedes aegypti and Anopheles gambiae constitute a branch in the phylogenetic tree (data not shown). With five large gaps (5-13 residues), the pro-regions of BmSpz1 and DmSpz1 are 14% identical and 35% similar to each other. The carboxyl-terminal active ligand is more conserved (identity: 20%; similarity: 45%). These include nine Cys residues that stabilize the structures via disulfide bond formation. Based on the sequence comparison (Fig. 1C) and published data [16,17], we suggest that pro-BmSpz1 contains four intrachain disulfide bridges: one in the pro-region and three in the ligand portion. The orphan Cys residue may form an interchain linkage with its counterpart on the other subunit. BmSpz1 contains one putative O-linked glycosylation site at Thr108 and two putative N-linked glycosylation sites at Asn75 and Asn220.
3.2. Spatial and temporal regulation of BmSpz1 transcription
We examined the mRNA levels of BmSpz1 in different tissues and developmental stages by RT-PCR. The BmSpz1 transcripts were most abundant in midgut, followed by fat body (Fig. 2A). The mRNA levels were much lower in silk gland, head, integument, ovary, testis, and eggs. The low constitutive transcription was detected in the total RNA samples extracted from whole larvae/pupae (Fig. 2B). Except for the third instar, BmSpz1 mRNA in all larval stages was less abundant than that in days 1-8 of the pupae.
Fig. 2.
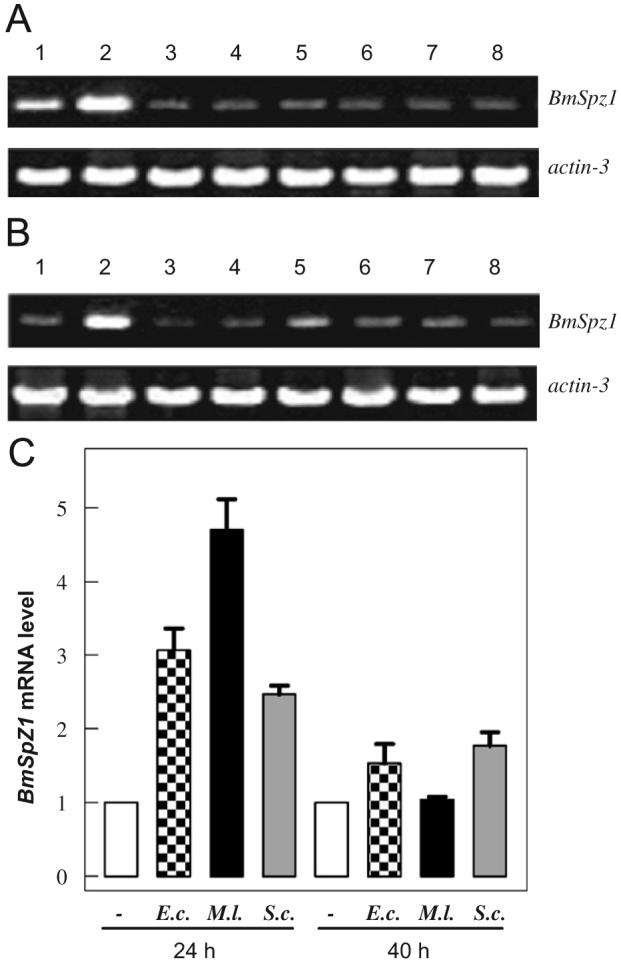
Inducibility, developmental, and tissue-specific expression of BmSpz1. (A) Tissue specificity. 1, fat body; 2, midgut; 3, silk gland; 4, head; 5, integument; 6, ovary; 7, testis; 8, egg. (B) Developmental profile. 1, 2nd instar; 2, 3rd instar; 3, 4th instar; 4, 5th instar; 5-8, day 1, 3, 5 and 8 of pupae. (C) Immune responsiveness. Day 3, 5th instar silkworm larvae were injected with water, E. coli, M. luteus, or S. cerevisiae cells (50 μl each). Fat body tissues were collected from the treated larvae 24 and 40 h later for total RNA extraction. Following RT-PCR and gel electrophoretic analysis, the relative band intensities were measured by densitometry and plotted on the bar graph as mean±SD (n = 3). The silkworm actin-3 was used as an internal standard to normalize the templates.
To test the hypothesis that BmSpz1 participates in immune responses, we compared the levels of BmSpz1 mRNA in fat body after the silkworm larvae were injected with water or different micro-organisms (Fig. 2C). There was a significant increase at 24 h after the immune challenge of M. luteus, E. coli, or S. cerevisiae. The transcript levels decreased and became more comparable to the control at 40 h, especially in the M. luteus-treated larvae. BmSpz1 seems to be involved in defense against bacterial and fungal infection, and its gene expression is tightly regulated.
3.3. Production and characterization of pro-BmSpz1 and BmSpz1 from E. coli
To explore its physiological roles, we cloned a BmSpz1 cDNA fragment and constructed two expression plasmids (pro-BmSpz1 /TevH6pQE60 and BmSpz1 /H6pQE60). E. coli JM109 cells harboring the plasmids expressed the full-length and truncated BmSpz1, which mimic the inactive proligand and active ligand. The insoluble recombinant proteins were extracted from purified inclusion bodies by 8 M urea and purified by affinity chromatography under the denaturing condition. After renaturation and concentration, we finally obtained 1.5 mg pro-BmSpz1 and 0.6 mg BmSpz1 from the E. coli cultures (11 each).
The purified pro-BmSpz1 and BmSpz1 migrated as tight doublets to 31 and 13 kDa positions on SDS-PAGE gel under reducing condition (Fig. 3A and B). Consistent with that, MALDI mass spectrometry of pro-BmSpz1 revealed two peaks at 30,973 ± 155 and 31,246 ± 156 Da. Similarly, two mass peaks were detected at 13,315 ± 27 and 13,613 ± 27 Da for the BmSpz1 sample. Since the calculated molecular masses of pro-BmSpz1 and BmSpz1 fusion proteins were 30,925 and 13,306 Da, respectively, the observed mass increases may have resulted from post-translational modification in E. coli. The mass fingerprint analysis of the pro-BmSpz1 doublet; however, failed to provide an explanation for such change (data not shown). Under non-reducing condition, most pro-BmSpz1 migrated to 31 kDa position and the rest of the protein was detected as diffused bands at around 60, 75, 120 and 150 kDa (Fig. 3C). BmSpz1 ran on the SDS-PAGE gel as a series of doublets at 22, 24, 33, 36, 44, 48, and 60-300 kDa positions. The 33 and 36 kDa bands, with intensities equal to or higher than the other doublets, may correspond to BmSpz1 dimers. Reexamination of the BmSpz1 mass spectrum revealed three small peaks at 26,645 ± 133, 26,942 ± 135 and 27,295 ± 136 Da, which are similar to the masses of the hetero- and homodimers of BmSpz1. These data suggest BmSpz1 is prone to oligomer formation and the dimers represent a significant portion of the total renatured protein.
Fig. 3.
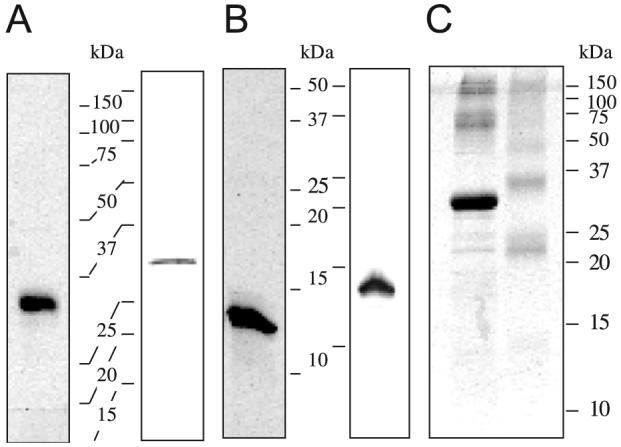
SDS-PAGE analysis of BmSpz1 and its precursor from E. coli. Affinity purified and renatured pro-BmSpz1 and BmSpz1 were treated with SDS sample buffer with or without β-mercaptoethanol, resolved on a 15% SDS-polyacrylamide gel by electrophoresis, and stained with Coomassie Brilliant Blue. Proteins on a duplicate gel were electrotransferred to a nitrocellulose membrane and subjected to immunoblot analysis using 1:2000 diluted antiserum against pro-BmSpz1 as the first antibody. (A) Reduced pro-BmSpz1: 1 μg for stained gel (left) and 50 ng for immunoblot analysis (right); (B) reduced BmSpz1: 1 μg for staining (left) and 100 ng for immunoblotting (right). and (C) unreduced pro-BmSpz1 (left, 0.5 μg) and BmSpz1 (right, 1 μg) on a stained SDS-PAGE gel.
3.4. Induction of antimicrobial peptide gene expression by BmSpz1
We tested whether or not BmSpz1 could mediate immune responses by stimulating antimicrobial peptide synthesis in the absence of infection. After injection of water, pro-BmSpz1 or BmSpz1 to the silkworm larvae, we dissected the fat body at 24 h for total RNA extraction. RT-PCR analysis indicated that transcription of eight antimicrobial peptide genes was significantly up-regulated by BmSpz1 (Fig. 4A). These genes are B. mori attacin-2, cecropin-A1, -B1, -D1, gloverin-A5, -B, lebocin-3, and moricin-A1. While low mRNA levels of several genes were detected in the negative control of water injection, no elevation in mRNA levels was observed after injection of pro-BmSpz1—the small increase in attacin-2 was comparable to another negative control of bovine serum albumin (data not shown). The major increase in transcript levels after BmSpz1 injection, therefore, was not caused by E. coli components that might have contaminated both pro-BmSpz1 and BmSpz1. The recombinant BmSpz1, which bypassed pathogen recognition and proteolytic activation, apparently bound to a toll-like receptor and induced immune gene expression.
Fig. 4.
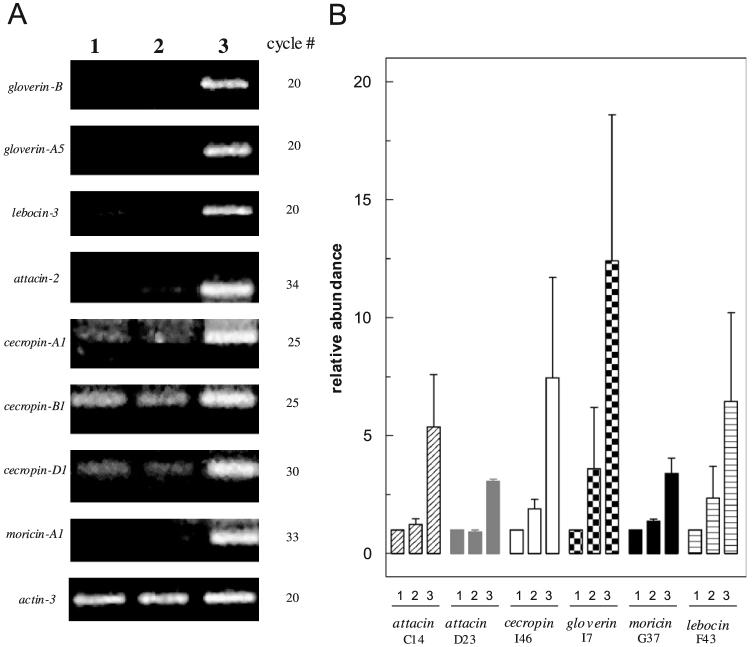
Induced expression of antimicrobial peptide genes in the silkworm and tobacco hornworm after BmSpz1 treatment. (A) B. mori. Total RNA was isolated from fat body 40 h after injection of water (1), pro-BmSpz1 (2), or BmSpz1 (3). cDNA samples from control and treated larvae were synthesized using reverse transcriptase, normalized with silkworm actin-3, and analyzed by PCR using primers specific for B. mori antimicrobial peptide genes. The PCR cycle numbers are indicated. (B) M. sexta. Fat body total RNA samples were isolated from M. sexta larvae 24 h after injection of water (1), pro-BmSpz1 (2), or BmSpz1 (3). The relative mRNA levels of antimicrobial peptides were examined by RT-PCR, electrophoresis, and densitometry. The relative band intensities (H2O = 1.0) were plotted on the bar graph as mean±SD (n = 3). cDNA samples were normalized with Manduca rpS3.
Could BmSpz1 up-regulate the transcription of antimicrobial peptide genes in other lepidopteran insects such as M. sexta? To answer this question, we did the same experiment and detected similar changes in the transcription of antimicrobial genes [18] in the tobacco hornworm (Fig. 4B). In this case, small elevations in M. sexta attacin (C14 and D23), cecropin (I46), gloverin (I7), moricin (G37) and lebocin (F43) transcript levels were detected after pro-BmSpz1 injection. Such changes were more prominent upon BmSpz1 treatment.
3.5. Cleavage of silkworm pro-BmSpz1 in M. sexta hemolymph
The finding that BmSpz1 regulates transcription of M. sexta immune genes led us to hypothesize that a similar signaling pathway may also exist in the tobacco hornworm, including spätzle processing enzyme, pro-spätzle and toll-like receptor. If so, could we use pro-BmSpz1 as a substrate to detect the M. sexta pro-spätzle activating proteinase? To test this approach, we incubated pro-BmSpz1 with M. luteus and plasma from immune challenged M. sexta larvae. Western blot analysis did show proteolytic processing but the cleavage was not very specific (Fig. 5A): at least 6 bands were detected in the range of 10-30 kDa. The faint band at ∼13 kDa may correspond to BmSpz1 which slightly induced the synthesis of M. sexta antimicrobial peptides (Fig. 4B). The proteolysis of pro-BmSpz1 was also observed using the silkworm plasma and M. luteus, but the smallest cleavage product was ∼15 kDa (Fig. 5B).
Fig. 5.
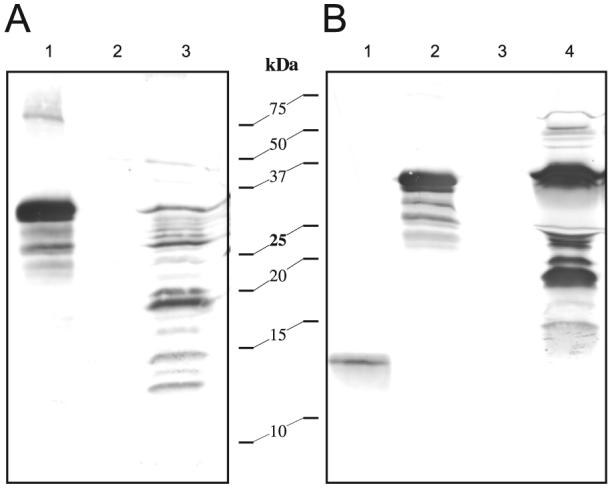
Cleavage of pro-BmSpz1 by proteinases in the plasma from M. sexta (A) and B. mori (B) larvae challenged by killed microbes. Pro-BmSpz1 (0.2 μg, 1 μl), induced plasma (2 μl), M. luteus (1 μg, 1 μl), and buffer (20 mM Tris, pH 8.0, 10 μl) were incubated at room temperature for 15 min. The reaction mixture (11 μl) were analyzed by 15% SDS-PAGE followed by immunoblotting using 1:2000 diluted antiserum to B. mori spätzle-1 as the first antibody. (A) Lane 1, pro-BmSpz1; lane 2, induced plasma; lane 3, pro-BmSpz1, induced plasma and M. luteus. (B) Lane 1, BmSpz1; lane 2, pro-BmSpz1; lane 3, induced plasma; lane 4, pro-BmSpz1, induced plasma and M. luteus.
4. Discussion
The interaction between Drosophila spätzle and toll receptor establishes the dorsoventral axis during embryonic development and induces the synthesis of antimicrobial peptides [1-3]. Although homologous genes for the extra- and intracellular pathway components have been identified in various insect genomes [19,20], it is not demonstrated that proteolytic activation of pro-spätzle plays a similar role in other insects. In this paper, we report the structure of BmSpz1 gene and the comparison of its gene product with Drosophila spätzle (Fig. 1). The conserved amino acid sequence and disulfide linkage pattern suggest that BmSpz1 and Drosophila spätzle may share a common tertiary structure. Nevertheless, we failed to isolate M. sexta spätzle cDNA using BmSpz1-specific probes (i.e., DNA and antibodies) under low stringent conditions (unpublished data).
Spatial and temporal regulation of BmSpz1 transcription, as well as induced expression upon injection of M. luteus or yeast, suggests that BmSpz1 participates in antimicrobial responses (Fig. 2). Injection experiment using the purified proteins further demonstrated that cleavage activation is required to generate BmSpz1 which induces the transcription of antimicrobial peptide genes (Fig. 4). For the first time, we provided biochemical evidence that pro-spätzle processing and toll pathway activation is an evolutionarily conserved strategy in a nondrosophiline insect. Much to our surprise, injection of BmSpz1 induced transcription of immunity-related genes in a different lepidopter-an insect.
Drosophila pro-spätzle and spätzle were both present in the form of dimers maintained by an interchain disulfide bond (16), and dimerization is critical for the biological activity of spätzle [17]. We detected a significant amount of dimers in the renatured BmSpz1 while pro-BmSpz1 mainly existed as monomers (Fig. 3C). Although these results agreed with the Drosophila model and activity data (Fig. 4), there were clear differences in the association states among the recombinant proteins. As Drosophila pro-spätzle and spätzle were produced in insect cells (16), we are now trying to express pro-BmSpz1 and BmSpz1 in a baculovirus-insect cell system and study its association. That may resolve some of the discrepancies observed in pro-spätzles and spätzles from the two insect species.
In vitro cleavage tests demonstrated that the recombinant pro-BmSPz1 was processed by B. mori and M. sexta plasma samples in the presence of M. luteus (Fig. 5). We did not detect any endogenous pro-BmSpz1, possibly because the glycoprotein was not recognized by antibodies raised against pro-BmSpz1 from E. coli. The detection of the ∼15 kDa faint band as the smallest cleavage product coincides with the result that pro-BmSpz1 injection did not induce immune gene transcription. The appearance of multiple cleavage products (including the ∼13 kDa faint band) after incubation with M. sexta plasma seems to be consistent with the slight increase of antimicrobial peptide gene expression after pro-BmSpz1 injection (Fig. 4).
In summary, we provided biochemical and molecular evidence for the possible existence of an extracellular proteinase cascade, a spätzle-like cytokine and its receptor, and an intracellular signaling pathway in lepidopteran insects, which lead to up-regulated transcription of immunity-related genes. We are planning to use pro-BmSpz1 as a substrate in the purification of M. sexta spätzle-processing enzyme.
Acknowledgments
This work was Rported by National Institutes of Health Grants GM58634 (to H.J.) and National Basic Research Program of China 2005CB121000 (to Z.X.). We thank Janet Rogers at OSU Recombinant DNA/Protein Resource Facility for assistance in peptide mass fingerprint analysis. We also thank Drs. Jack Dillwith and Junpeng Deng for their helpful comments on the manuscript. This article was approved for publication by the Director of Oklahoma Agricultural Experimental Station and Rported in part under Project OKLO2450.
Abbreviations
- BmSpz1 and pro-BmSpz1
Bombyx mori spätzle-1 and its precursor
- MALDI-TOF
matrix-assisted laser desorption ionization-time of flight
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- RT-PCR
reverse transcriptase-polymerase chain reaction
References
- [1].Morisato D, Anderson KV. Signaling pathways that establish the dorsal-ventral pattern of Drosophila embryo. Ann Rev Genet. 1995;29:371–99. doi: 10.1146/annurev.ge.29.120195.002103. [DOI] [PubMed] [Google Scholar]
- [2].Wang L, Ligoxygakis P. Pathogen recognition and signaling in the Drosophila innate immune response. Immunobiology. 2006;211(4):251–61. doi: 10.1016/j.imbio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- [3].Moussian B, Roth S. Dorsoventral axis formation in the Drosophila embryo—shaping and transducing a morphogen gradient. Curr Biol. 2005;15(21):R887–99. doi: 10.1016/j.cub.2005.10.026. [DOI] [PubMed] [Google Scholar]
- [4].Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart JM. Activation of Drosophila toll during fungal infection by a blood serine protease. Science. 2002;297(5578):114–6. doi: 10.1126/science.1072391. [DOI] [PubMed] [Google Scholar]
- [5].Jang IH, Chosa N, Kim SH, Nam HJ, Lemaitre B, Ochiai M, et al. A spätzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev Cell. 2006;10(1):45–55. doi: 10.1016/j.devcel.2005.11.013. [DOI] [PubMed] [Google Scholar]
- [6].Mulinari S, Hacker U, Castillejo-Lopez C. Expression and regulation of spätzle-processing enzyme in Drosophila. FEBS Lett. 2006;580(22):5406–10. doi: 10.1016/j.febslet.2006.09.009. [DOI] [PubMed] [Google Scholar]
- [7].Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, et al. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for toll activation. Curr Biol. 2006;16(8):808–13. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- [8].Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306(5703):1937–40. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- [9].Dunn P, Drake D. Fate of bacteria injected into naïve and immunized larvae of the tobacco hornworm, M. sexta. J Invert Pathol. 1983;41:77–85. [Google Scholar]
- [10].Cheng T, Zhao P, Liu C, Xu P, Gao Z, Xia Q, et al. Structures, regulatory regions, and inductive expression patterns of antimicrobial peptide genes in the silkworm Bombyx mori. Genomics. 2006;87(3):356–65. doi: 10.1016/j.ygeno.2005.11.018. [DOI] [PubMed] [Google Scholar]
- [11].Lee E, Linder ME, Gilman A. Expression of G-protein alpha subunits in E. coli. Meth Enzymol. 1994;237:146–64. doi: 10.1016/s0076-6879(94)37059-1. [DOI] [PubMed] [Google Scholar]
- [12].Jiang H, Wang Y, Yu XQ, Kanost MR. Prophenoloxidase-activating proteinase-2 from hemolymph of M. sexta. A bacteria-inducible serine proteinase containing two clip domains. J Biol Chem. 2003;278(6):3552–61. doi: 10.1074/jbc.M205743200. [DOI] [PubMed] [Google Scholar]
- [13].Wang Y, Jiang H. Prophenoloxidase (proPO) activation in M. sexta: an analysis of molecular interactions among proPO, proPO-activating proteinase-3, and a cofactor. Insect Biochem Mol Biol. 2004;34(8):731–42. doi: 10.1016/j.ibmb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- [14].Cherbas L, Cherbas P. The arthropod initiator: the capsite consensus plays an important role in transcription. Insect Biochem Mol Biol. 1993;23(1):81–90. doi: 10.1016/0965-1748(93)90085-7. [DOI] [PubMed] [Google Scholar]
- [15].Hultmark D. Immune reactions in Drosophila and other insects: a model for innate immunity. Trends Genet. 1993;9(5):178–83. doi: 10.1016/0168-9525(93)90165-e. [DOI] [PubMed] [Google Scholar]
- [16].DeLotto Y, DeLotto R. Proteolytic processing of the Drosophila spätzle protein by easter generates a dimeric NGF-like molecule with ventralizing activity. Mech Dev. 1998;72(12):141–8. doi: 10.1016/s0925-4773(98)00024-0. [DOI] [PubMed] [Google Scholar]
- [17].Weber AN, Moncrieffe MC, Gangloff M, Imler JL, Gay NJ. Ligand-receptor and receptor-receptor interactions act in concert to activate signaling in the Drosophila Toll pathway. J Biol Chem. 2005;280(24):22793–9. doi: 10.1074/jbc.M502074200. [DOI] [PubMed] [Google Scholar]
- [18].Zhu Y, Johnson TJ, Myers AA, Kanost MR. Identification by subtractive Rpression hybridization of bacteria-induced genes expressed in M. sexta fat body. Insect Biochem Mol Biol. 2003;33(5):541–59. doi: 10.1016/s0965-1748(03)00028-6. [DOI] [PubMed] [Google Scholar]
- [19].Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298(5591):159–65. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- [20].Evans J, Aronstein K, Chen YP, Hetru C, Imler JL, Jiang H, et al. Immune pathways and defense mechanisms in the honey bee, Apis mellifera. Insect Mol Biol. 2006;15(5):645–56. doi: 10.1111/j.1365-2583.2006.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]