Abstract
The Tolloid metalloproteases are pleiotropic enzymes that are important for many developmental processes. This study describes the isolation and characterization of a novel Tolloid family member from the pond turtle Pseudemys scripta elegans. The turtle Tolloid, designated tTll, is found in two forms. The first, tTlls, contains a signal sequence which may provide a mechanism for secretion. The second, tTllc, does not contain a signal sequence and is likely cytoplasmic. Sequence analysis of tTll revealed that it is most closely related to chicken Tll-2 although tTll domain structure is different. We examined the expression of tTll mRNA by real time RT-PCR and found the highest expression in the cerebellum with lower levels in the brain stem and cortex. This expression pattern is similar to the expression of the Tolloid mouse orthologues Tll-1 and Tll-2 with highest levels of expression in the cerebellum and lower levels in the brain stem and cortex.
Section: Cellular and Molecular Biology of Nervous System
Keywords: Tolloid, Turtle, PCR, Gene Expression
1. Introduction
The Tolloid family of metalloproteases plays many important roles in embryonic patterning and tissue morphogenesis during embryogenesis of both vertebrates and invertebrates (Mullins 1998; Yamamoto and Oelgeschlager 2004). In the fruit fly, Drosophila melanogaster, Tolloid is required for dorsal ventral patterning. Tolloid potentiates the activity of decapentaplegic (DPP), a transforming growth factor-β (TGF-β) family member, by antagonizing short-gastrulation (SOG), a protein that binds DPP and prevents it from binding to its receptor. Tolloid and DPP are expressed by cells on the dorsal aspect of the embryo, whereas SOG is expressed on the ventral side of the embryo (Ferguson and Anderson 1992). This results in a gradient of DPP from the dorsal to ventral aspect of the embryo that is responsible for patterning.
In vertebrates Tolloid proteins play important roles in embryogenesis as well as in adult tissues. In mammals, there are three homologues of Drosophila Tolloid; Tolloid/BMP-1, Tolloid-like 1 (Tll-1) and Tolloid-like 2 (Tll-2). The Tolloid/BMP-1 gene produces two alternatively spliced transcripts, one encoding BMP-1 and the other encoding Tolloid, whereas TLL-1 and TLL-2 genes produce single transcripts (Takahara et al. 1994; Clark et al. 1999; Scott et al. 1999). Both BMP-1 and Tll-1 are capable of cleaving chordin, the mammalian homologue of Drosophila SOG (Scott et al. 1999). The Tolloid family of metalloproteases also processes a number of proteins that comprise the extracellular matrix including procollagen, laminin, and biglycan. In mice, knockouts have been made for both the Tolloid/BMP-1 and TLL-1 genes. The Tolloid/BMP-1 knockout results in perinatal lethality and a lack of processing of fibrillar collagen and laminin (Suzuki et al. 1996). Interestingly, the TLL-1 knockout results in embryonic lethality due to cardiac defects (Clark et al. 1999). When double knockouts were made for both the Tolloid/BMP-1 and TLL-1 genes the embryos died in utero at the same gestational age as the TLL-1 knockouts. However, there appeared to be an additional neural phenotype. The double knockouts showed a reduced number of cells within the developing neural system when compared with wild-type or single knockout mice (Clark, unpublished data). Further examination of the expression pattern of TLL-1 in normal mice revealed Tll-1 mRNA in the hippocampus in a punctate pattern (Scott et al. 2000). Characterization of Tll-1 mRNA expression showed that the number of positive cells were increased in the hippocampus of mice with access to running wheels, conditions that increase neurogenesis, and were reduced in the hippocampus of mice under conditions that decrease neurogenesis (Tamura et al. 2005). Since mice with increased neurogenesis show improved learning and memory (Gould and Tanapat 1999) it is possible that Tll-1 plays a role in this process. This role is further supported by studies in the marine mollusk Aplysia in which a Tolloid-like gene (apTolloid/BMP-1) was found to be regulated during a nonassociative learning paradigm that induced long-term sensitizaton (Liu et al. 1997).
The present study is a first step in defining a role for the Tolloid-like genes in mechanisms of plasticity such as neurogenesis and learning. One model system we have examined extensively is the classically conditioned abducens nerve response generated in an isolated brain stem preparation from turtles (Keifer 2003; Keifer and Clark 2003; Mokin et al. 2006). This model is an in vitro neural correlate of eyeblink classical conditioning that demonstrates many of the hallmarks of conditioning in mammals. Given the advantage of this preparation for studies of cellular mechanisms underlying associative learning, we isolated the homolog of mammalian Tolloid in pond turtles. This will allow studies to identify the role of Tolloid in learning and memory to be performed in this system. The following work describes our characterization of the reptilian homologue of mammalian Tolloid including a comparison of its structure and expression with mammalian Tll-1 and Tll-2.
2. Results
Isolation of turtle Tolloid-like 1
To identify a turtle orthologue of mammalian Tolloid-like 1, we designed degenerate primers based on the sequence of the chicken, mouse zebrafish and frog Tll-1 sequences and performed reverse transcription-polymerase chain reaction (RT-PCR). Following RT-PCR we initially identified a 1080 bp fragment that was sequenced. From the resulting sequence we designed primers and performed both 5′- and 3′-RACE to identify the 5′ and 3′ termini of the presumptive tTll mRNA. We identified two transcripts, tTlls and tTllc, which differ at the 5′-end (Fig. 1). Both transcripts contain open reading frames flanked by 5′- and 3′-untranslated sequences. The 3′-untranslated regions of both tTlls and tTllc appear to be full length since they both terminate in a poly(A) tail preceded, 29 bases upstream, by the sequence AGTAAA, which has previously been shown to function as a polyadenylation signal (Beaudoing et al. 2000). BLAST searches were used to identify sequence identity to other sequences in the GenBank database. The tTll sequences show high similarity at the nucleotide level, to the predicted Gallus gallus gene similar to TLL-2 (87% identical), Homo sapiens TLL-2 (83% identical), Bos taurus TLL-2 (83% identical), Mus musculus TLL-2 (82% identical) and to the other known vertebrate TLL-2 proteins.
Figure 1.
cDNA Sequences of turtle tolloid-like secreted (tTLLs) and turtle tolloid-like cellular (tTLLc). tTLLs sequence is shown complete. Alignment was performed using the MegAlign program (DNASTAR). tTLLc is shown only where it differs from tTLLs. Differences between sequences are shown by highlighting. Fragments that were sequenced again to confirm existing of both tTLLs and tTLLc variants shown by bold font, sites for primer are underlined (Table 2). Sites for RACE and for RT-PCR are shown by bold font and arrows. Regions that overlap are highlighted. Identical sequences are boxed and the polyadenylation signal is circled. The ATG start and TGA stop codons are underlined. The sites for primers and the probe used for Real Time RT-PCR are also shown.
The tTll s and tTll c conceptual translation products
Analysis of the conceptual translation products of tTlls and tTllc reveal a 2814 nucleotide transcript for tTllc that encodes an 838 amino acid protein and a 2595 nucleotide transcript that encodes an 801 amino acid protein in tTlls. The difference between tTlls and tTllc occurs at the 5′ end of the transcript and influences the predicted cellular location of the protein product. Analysis of the sequence using SignalP 3.0 Server indicates that tTlls contains a signal sequence that is predicted to specify the protein for translation into the endoplasmic reticulum, presumably to be secreted similar to other members of the Tolloid family. In contrast, tTllc does not contain a signal sequence and is predicted to be found only in the cytoplasm (Fig. 2).
Figure 2.
Alignment of the amino acid residues of human mTll-2 (hmTll-2) (NCBI Gen Bank accessions CAH72234) and turtles tTllc and tTlls. Black, dash and gray lines show metalloprotease, SUB and EGF domains respectively. Alignment was performed using the MegAlign program (DNASTAR). The metalloendopeptidase active site motif abXHEbbHbc (Rawlings and Barrett 1993) is enclosed by a dashed box. The signal peptide cleavage site was predicted as described by Nielsen (Bendtsen et al. 2004), using SignalP 3.0 Server.
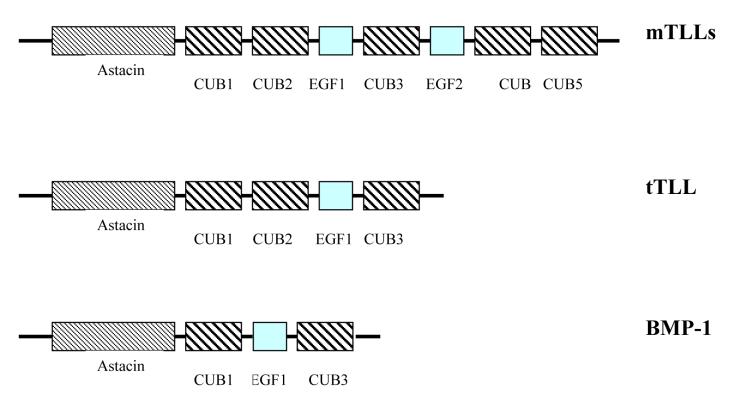
Comparison of the amino acid sequences of tTll with proteins from the GenBank database revealed that tTll is most similar to mammalian Tll-2 from Bos taurus (91% similar, 84% identical), Homo sapiens Tll-2(90% similar, 84% identical), and Mus musculus Tll-2 (89% similar, 82% identical). tTll also shows a high degree of similarity in domain architectures when compared to mammalian Tll-1 and Tll-2, except that tTll is markedly smaller relative to the mammalian Tolloid-like genes. tTllc and tTlls include one astacin-like protease domain, three CUB protein-protein interaction domains, and only one EGF motif (Bond and Beynon 1995), instead of five CUB domains and two EGF domains found in mammalian Tolloids (Fig. 3). Although tTll is most similar to mTll-2 its domain structure most closely resembles that of mammalian BMP-1.
Figure 3.
Scheme of domain architectures of known vertebrate TLL proteins (TLL-1, TLL-2), domain architectures of tTLLs proteins and bone morphogenetic protein 1 (BMP-1). Analysis of domain architecture was performed using Conserved Domain Architecture at NCBI.
The protease domain of tTll is virtually identical to that of human Tll-2. There are only 5 differences out of 193 amino acids within the domain. Of the known astacin-metalloproteases, approximately half contain an HEXXH motif, which has been shown in crystallographic studies to form part of the metal-binding site. The HEXXH motif is common, but can be more stringently defined for metalloproteases as abXHEbbHbc, where ‘a’ is most often valine or threonine, ‘b’ is an uncharged residue, and ‘c’ a hydrophobic residue (Rawlings and Barrett 1995). The tTll metal binding site is identical to that found in human Tll-2 (Fig. 2).
Isolation of a partial sequence of turtle actin
In order to quantify relative levels of tTll from different brain regions we needed a constitutively expressed turtle gene as a loading control. Since few turtle genes have been described we isolated, by RT-PCR, a partial sequence of turtle actin. Degenerate primers were designed based on the sequence of chicken, mouse, zebrafish and frog actin sequences. We isolated two different 800 bp cDNA fragments which, following BLAST searches, were identified as turtle β-, and γ-actin (data not shown).
Relative levels of tTlls and tTllc in turtle brain
To determine if tTlls and tTllc mRNAs are both expressed in the turtle brain, we performed RT-PCR and compared the expression to that of turtle actin. Total RNA from turtle cerebellum was used to synthesize cDNA after which PCR was performed to identify tTlls, tTllc and turtle actin using the two sets of primers that resulted in two different size products for each transcript (Table 1). Figure 4 shows that both tTlls (Lanes 2 and 4) and tTllc (Lanes 6 and 8) are expressed in turtle cerebellum with slightly higher expression of tTllc than tTlls. This suggests that both isoforms of tTll are expressed within the turtle cerebellum.
Table 1.
Primers used to confirm the existence of both variants of tTLL.
| Target | Forward primer | Reverse primer | Size of PCR products (bp). |
|---|---|---|---|
| actin | tgaccgagcgtggctacag | ctttacggatgtcaacgtcacact | 296 |
| tTLLs | gggaccctgctgctgctacttc | caagtcctcttcgtctaaagcaatatc | 163 |
| tTLLs | gggaccctgctgctgctacttc | gcgttcctcctgccatccttttt | 326 |
| tTLLc | gtcccccaggctacccatat | gttctgtctatctgaaacagcttcaag | 273 |
| tTLLc | gtcccccaggctacccatat | gcgttcctcctgccatccttttt | 416 |
Figure 4.
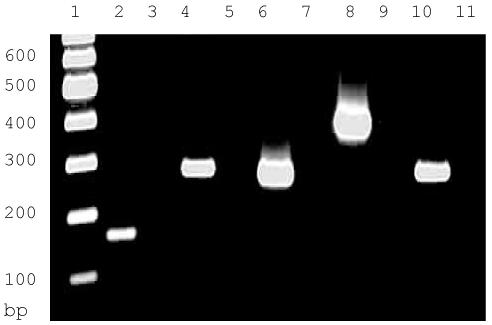
Relative levels of tTlls and tTllc in the cerebellum of the turtle as determined by RT-PCR. Lane 1, DNA ladder; Lane 2, tTlls 163 bp reaction; Lane 3, control tTlls 163 bp reaction; Lane 4, tTlls 326 bp reaction; Lane 5, control tTlls 326 bp reaction; Lane 6, tTllc 273 bp reaction; Lane 7, control tTllc 273 bp reaction; Lane 8, tTllc 416 bp reaction; Lane 9, control tTllc 416 bp reaction; Lane 10, turtle actin (296 bp); Lane 11, control turtle actin.
Expression of tTlls in turtle brain
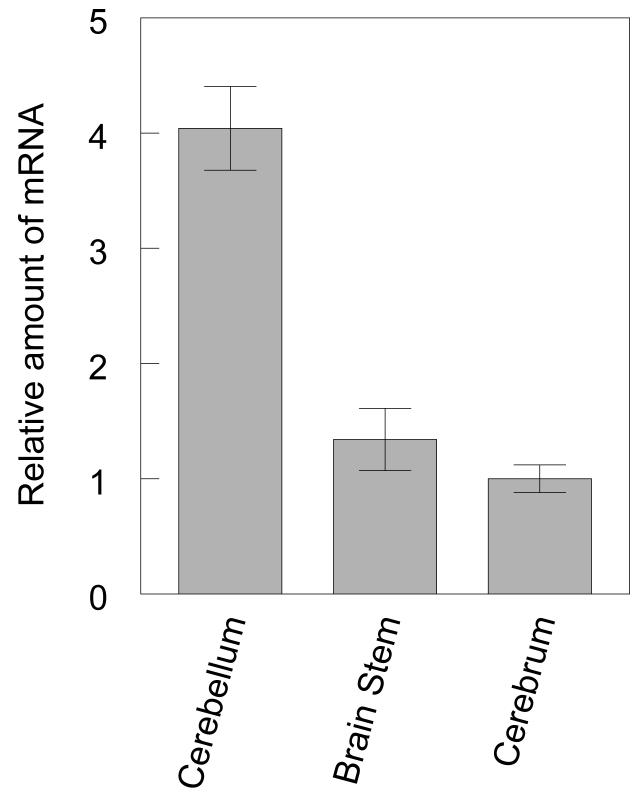
To further characterize the expression of tTll and confirm our PCR results, we examined tTll mRNA expression by Northern blot analysis. The results of the Northern blot showed a single tTll mRNA species with an approximate size of 2.8 kb when compared with the 18S and 28S ribosomal subunits. The length of the tTll mRNA correlates well with size of the tTlls and tTllc cDNAs that we have isolated, which are predicted to be 2.6 and 2.8 nucleotides, respectively (Fig. 5). We quantified the relative levels of the mRNA by densitometry using Image Quant version 5.2 (BioRad) and normalized the results to turtle actin. tTll expression in the turtle cerebellum is two-fold higher than in the brain stem and cerebrum.
Figure 5.
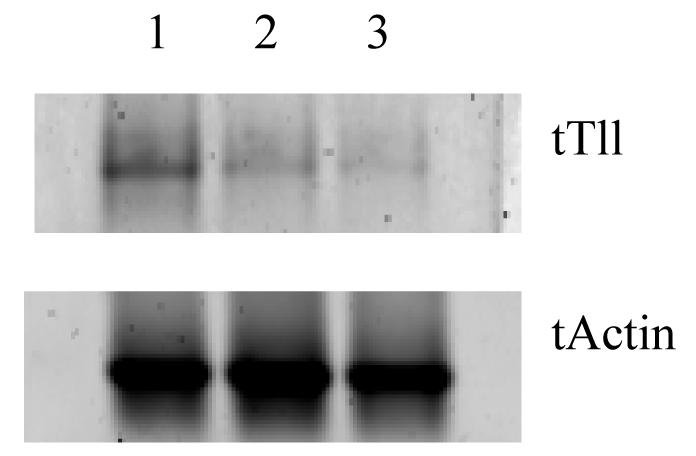
Northern blot analysis of tTll in the three regions of the turtle brain. Lane 1, Cerebellum; Lane 2, Brain stem; Lane 3, Cerebrum.
Comparison of tTll in turtle brain with Tll-1 and Tll-2 in mouse brain
We employed the use of Real Time RT-PCR to compare the level of expression of tTll in turtle brain with Tll-1 and Tll-2 in mouse brain. The primers and probes for real time RT-PCR of tTll, turtle actin, Tll-1, and Tll-2 mRNA are shown in Table 2. Real time RT-PCR of tTll revealed the highest expression in the cerebellum (Fig. 6a). tTll was also expressed within the brain stem and cerebrum but the expression was over two-fold lower than in the cerebellum, confirming the results of our Northern blot analysis.
Table 2.
Primers and probes used for the analysis of turtle and mammalian Tll by real time PCR.
| Target | Species | Forward primer | Reverse primer | MGB probe |
|---|---|---|---|---|
| Tll1 | mouse | cgcccagaccgagacaac | gtactcttgacctggctggatgtt | atgtcaccatcattagag |
| Tll2 | mouse | gatggcatgagcccagtgt | cctccatgcccagtagtcaga | ctggtgacttggctgc |
| GAPDH | mouse | gggaagcccatcaccatctt | cggcctcaccccatttg | agcgagaccccactaa |
| tTll | turtle | aggcacgacagttgtgcttatg | caatcaaagggctgtcctcagt | ctacttggaaatccgagacggccc |
| t actin | turtle | agggaaatcgtgcgtgacat | gcggcagtggccatctc | aagctgtgctatgttgctctagacttc |
Figure 6.
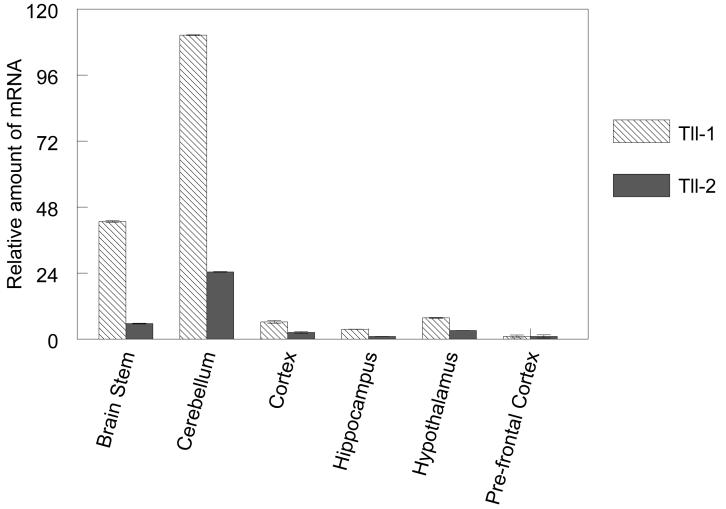
Comparison of Tll expression in different regions of the turtle brain and mouse brain by real time RT-PCR. A. Histogram shows the relative levels of tTll in the turtle brain. B. Relative levels of expression of Tll-1 and Tll-2 in different regions of the mouse brain. Error bars indicate standard deviation.
Both mammalian Tll-1 and Tll-2 are widely expressed in the adult mouse brain. Real time RT-PCR shows that, as in the turtle, Tll-1 and Tll-2 are expressed at the highest levels in the cerebellum (Fig. 6b). The brain stem also expresses both Tll-1 and Tll-2 at relatively high levels, although approximately 2-fold lower than the cerebellum. The cortex, hippocampus and hypothalamus also express Tll-1 and Tll-2 at similar levels, whereas the prefrontal cortex shows the lowest expression of all regions examined. Interestingly, although Tll-2 is expressed at significantly lower levels than Tll-1, the relative levels of Tll-2 among the different regions mimic the expression of Tll-1 closely.
In situ analysis of tTll expression
In situ hybridization revealed that tTll expression was found throughout the brainstem and cortex. High levels of tTll expression were observed in the cerebellum which confirms our real time-PCR results (Figure 7a) and is similar to the expression of mammalian Tll-1 that we have previously reported (Scott et al. 2000). More specifically, tTll expression is particularly enhanced in the granule cell layer of the cerebellar cortex compared to either the Purkinje cell layer or the molecular layer (Fig. 7 A-B). However, tTll expression was observed in a subpopulation of Purkinje cells scattered throughout the cerebellar cortex (data not shown). No label for tTll was observed in cerebella of controls (Fig. 7 C-D). Additionally, distinct regions of punctate expression of tTll appeared in the molecular layer of the cerebellum but not in controls (Figure 7 E-H, arrows). These clusters of intense label just superficial to the Purkinje cell layer may be basket cells or regions of tTll aggregates near the proximal dendrites of Purkinje cells.
Figure 7.
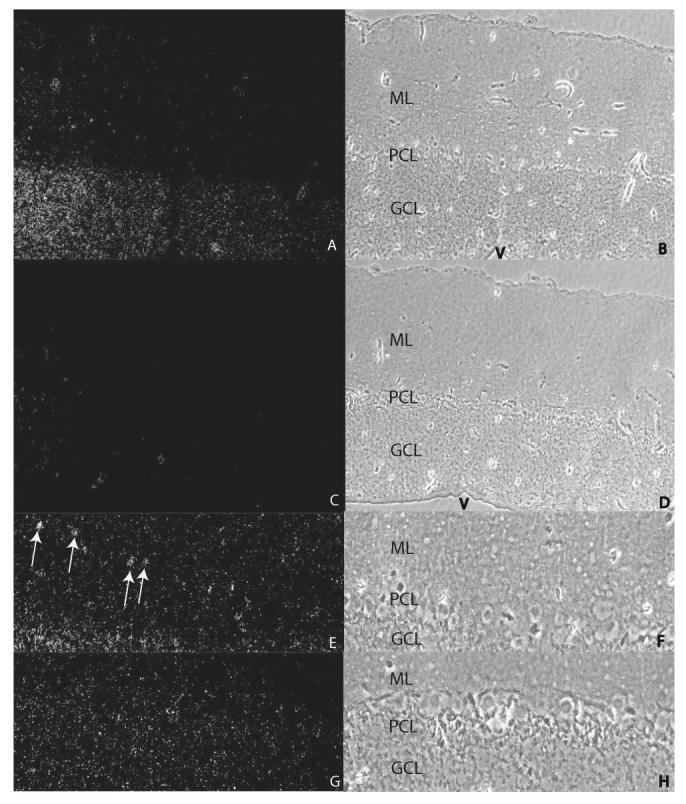
In situ hybridization of tTll in turtle cerebellum. Darkfield (100X; A) and phase contrast (B) images of turtle cerebellum hybridized with antisense tTll probe. There is dense label in the GCL compared to the PCL and the ML. Darkfield (100X; C) and phase contrast (D) images of turtle cerebellum hybridized with sense tTll probe show no label. Darkfield (200X; E) and phase constrast (F) images of turtle cerebellum hybridized with antisense tTll probe. Darkfield (200X; G) and phase contrast (H) images of turtle cerebellum hybridized with sense tTll probe. The arows in E indicate regions of the ML that contain clusters of tTll label. ML, molecular layer; PCL, Purkinje cell layer; GCL, granule cell layer; V, ventricle.
3. Discussion
We have isolated the one of the turtle orthologues of the mammalian Tolloids. Our description of tTll compliments previous studies that have described the Tolloid family of genes in a number of vertebrate species that includes birds and now reptiles. We have identified two transcripts, tTlls and tTllc, which correspond to proteins that are predicted to be secreted and cytosolic, respectively. tTlls contains a signal peptide and is predicted to have an unmodified molecular weight of 111 kDa. tTllc does not possess a signal peptide but contains 37 more amino acid residues than tTlls resulting in a predicted molecular weight of 115 kDa. Since tTlls and tTllc contain identical sequences except for the 5′ end, we speculate that these transcripts are the products of alternative splicing of the same primary transcript. Alternative splicing has also been shown for mouse and human Tolloid (Takahara et al. 1994). The mammalian Tolloid gene produces two alternatively spliced transcripts that encode BMP-1 as well as Tolloid. The resulting proteins have different domain structures as well as substrate specificities (Scott et al. 1999).
The function of Tolloid expression beyond pattern formation in adult animals is presently poorly understood. In mammals, Tll-1 and Tll-2 have their highest levels of expression in brain in the cerebellum, and lesser amounts are expressed in the brain stem and regions of the cerebrum. A similar distribution of mRNA is also true for tTll (Takahara et al. 1996). Why levels of expression are substantially higher in the cerebellum compared to other brain regions is unclear but this pattern appears to be conserved in vertebrates. Evidence suggests that the Tolloids facilitate the activity of neurotrophic factors, such as the BMPs, and may function in regulation of neurogenesis and mechanisms of synaptic plasticity. Tamura et al. (2005) found a significantly greater number of Tll-1 expressing cells in the hippocampus of mice provided access to running wheels than in non-running controls, conditions in which neurogenesis is enhanced (van Praag et al. 1999; Tamura et al. 2005). Moreover, they described novel glucocorticoid response elements within the Tll-1 promotor and that Tll-1 expression is negatively regulated by glucocortcoids. Stress and glucocorticoid levels also negatively affect neurogenesis. It was hypothesized that one function of Tll-1 may be to potentiate BMP signaling and therefore hippocampal neurogenesis. A Tolloid-like gene has also been shown to be regulated during non-associative learning in Aplysia (Liu et al. 1997). This raises the intriguing possibility that Tolloids may have a role in mechanisms of synaptic plasticity as well as neurogenesis. In turtles, we have extensively studied brain stem preparations that exhibit a form of eyeblink classical conditioning entirely in vitro (Keifer 2003; Keifer and Clark 2003). This learning occurs quickly, within hours, and is robust. In addition to postsynaptic mechanisms involving trafficking of AMPA-type glutamate receptors to synapses during the learning, we have also identified the involvement of coordinate presynaptic mechanisms in the form of significantly enhanced levels of the presynaptic protein synaptophysin (Mokin and Keifer 2004). Preliminary data from RT-PCR studies indicate that tTll is significantly upregulated during conditioning as compared to pseudoconditioned controls (Keifer et al. 2006). Future studies will be aimed at examination of a potential role for the Tolloid-like genes, specifically tTll, in the regulation of in vitro classical conditioning using this model system.
Conclusions
We have identified a reptilian orthologue of the Tolloid gene family, tTll, in the turtle. There are two transcripts of tTll, one that appears to encode a secreted form of the protein (tTlls) and the other that appears to encode a cytosolic form (tTllc). The domain structure of tTll most closely resembles that of mammalian BMP-1 whereas the sequence is most similar to Tll-2. The expression of tTll closely mirrors that of mammalian Tll-1 and Tll-2 in the brain. Our real time RT-PCR assays show that tTll is expressed at the highest levels in the cerebellum followed by the brain stem and cerebrum. These results will provide the means to study the role of Tolloids in learning and memory in a reptile model.
4. Experimental procedures
Reverse transcriptase-PCR (RT-PCR)
Total RNA was isolated from the brain stem, cerebellum and telencephalon of the pond turtle Pseudemys scripta elegans using the MELT Total RNA Isolation System (Ambion, Austin, TX). The RNA was reverse transcribed with the Strata Script One-Tube RT-PCR System (Stratagene, La Jolla, CA) according to the manufacturer’s instructions. Degenerate primers were designed for conserved regions of Tolloid-like 1 from Gallus gallus, Xenopus laevis, Danio rerio, and Mus musculus. PCR was carried out using the following primers: forward primer (5′-TATGAYTTTGACAGYATCATGCATATGCC-3′) and reverse primer (5′-GGRGCWACYACYTGCCAMACACAGTTTTRT-3′). An additional set of degenerate primers was designed for the conserved region of actin using the known sequences from Mus musculus, Xenopus tropicalis, Gallus gallus, and Danio rerio. PCR was carried out using the following primers: forward primer (5′-GAYATGGARAAGATYTG GCAYCAYMS-3′) and reverse primer (5′-YTTDSTRATC CACATYTG YTGRAAGGT-3′). PCR fragments were purified from agarose gels with Zymoclean Gel DNA Recovery kit (Zymo Research, Orange, CA), cloned into pCR2.1 (Invitrogen, Carlsbad, CA) according to instructions from the manufacturer and sequenced (Iowa State University Sequencing Facility, Ames, IA).
Identification of the 5′ and 3′ termini of tTll
Fragments containing the ends of turtle Tolloid homolog (tTll) cDNA were amplified using a FirstChoice RLM-RACE kit (Ambion), according to the manufacturer’s protocol. We used nested primers 5′-CCAGTAGCCATCACGCACCTCCACAT-3′ and 5′-AGTTGGCCGAACCCCATTGTCATCTC-3′ for 5′RACE. For 3′RACE we used nested primers 5′-CGGCCAGACAACGGTGGGTGTGA-3′ and 5′-CCTGTGAACCTGGCTACG-3′. Products of the RACE reactions were cloned into pCR2.1 and sequenced as described above.
Sequence analysis
The sequence of tTll and turtle actin (t actin) were analyzed for similarities with known sequences using the Blast tool and the GenBank sequence database (Altschul et al. 1990). Multiple alignments were performed with the MegAlign tool of LaserGene 6 (DNAstar, Madison, WI).
Northern blot hybridization
Total RNA from the turtle cerebellum, brain stem and cerebrum was isolated using TRI reagent (Molecular Research Center, Inc.) according to the manufacturer’s protocol. 35 μg of total RNA was separated by electrophoresis through a formaldehyde gel. The RNA was transferred and fixed on Hybond-N+. Probe preparation was performed using the Primer-it II random primer labeling kit (Stratagene).
A 1021 bp fragment corresponding to the region from bases 889 through 1909 relative to the start codon of tTlls was used as the probe (this region corresponds to bases 1000 through 2020 in tTllc). This region contains the CUB1, CUB2 and part of EGF1 domains, but not the protease domain. The complete actin PCR product isolated from turtle was used as a template for probe preparation as described above. Hybridization was performed using MiracleHyb hybridization solution (Stratagene) according to the manufacturer’s protocol. Following hybridization, the blot was visualized by autoradiography.
Real time RT-PCR
Real time RT-PCR was performed using 50 ng total RNA per reaction. RNA was combined with primer/probe sets and TaqMan® Gold RT-PCR Master Mix (Applied Biosystems, Inc., Foster City, CA). Gene-specific primers and probes were created for Pseudemys scripta elegans tTll, t actin and mouse Tll-1, Tll-2, and glyceraldehyde phosphate dehydrogenase (GAPDH) using the Primer Express Software (Applied Biosystems, Inc.) as shown in Table 2. Real time assays were run on an ABI 7000 (Applied Biosystems, Inc.). The real-time PCR profile consisted of one cycle at 48°C for 30 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. All reactions were repeated twice. The results of real-time PCR of mouse Tll-1 and Tll-2 were normalized to GAPDH. Turtle Tll expression was normalized to t actin.
Conventional RT-PCR
To determine if both variants of tTll were expressed in the turtle, we used primers designed to distinguish between tTlls and tTllc. To identify tTlls the primer set was 5′-GGGACCCTGCTGCTGCTACTTC-3′ and 5′-CAAGTCCTCTTCGTCTAAAGCAATATC-3′ for the 163 bp product and 5′-GGGACCCTGCTGCTGCTATTC-3′ and 5′-GCGTTCCTCCTGCCATCCTTTTT-3′ for the 326 bp product (Table 1). To identify tTllc the primer set was 5′-GTCCCCCAGGCTACCCATAT-3′ and 5′-GTTCTGTCTATCTGAAACAGCTTCAAG-3′ for the 273 bp product and 5′-GTCCCCCAGGCTACCCATAT-3′ and 5′-GCGTTCCTCCTGCCATCCTTTTT-3′ for the 416 bp product. 100 ng of total RNA was reverse transcribed with the Strata Script One-Tube RT-PCR System (Stratagene) according to the manufacturer’s instructions. For control reactions the reverse transcriptase was omitted. The RT-PCR consisted of one cycle at 42°C for 40 min and 94°C for 1 min, followed by 30 cycles at 94°C for 40 s, 50°C for 30 s, and 72°C 40 s. The resultant fragments were separated on a 2% agarose gel and stained with ethidium bromide. The 326 bp tTlls product and the 416 bp tTllc product were purified from agarose gels (Zymoclean Gel DNA Recovery Kit, Zymo Research), cloned into the pCR2.1 plasmid (Invitrogen) and sequenced as described.
In situ hybridization
In situ hybridization was performed as described in Tamura et al., 2005. Briefly, turtle brainstem preparations were fixed overnight in 4% paraformaldehyde, dehydrated in graded ethanols, and embedded in paraffin. Five micron sections were mounted on gelatin coated slides. The sections were rehydrated in graded ethanols and hybridized to 1021 base 35S-labeled probes corresponding to nucleotides 889-1909 of tTlls. This probe will hybridize to both forms of tTll. The probes were synthesized using the Maxiscript (Ambion) kit following the manufacturer’s protocol. Following post-hybridization washes the slides were dehydrated and exposed to photographic emulsion (Amersham) for 10 days.
Acknowlegments
Supported by NIH grant P20 RR015567, which is designated as a Center of Biomedical Research Excellence (COBRE) to J.K. and T.G.C., NIH grant NS051187 to J.K., and AHA Northland Affiliate Scientist Development Grant 0235534Z to T.G.C.
Abbreviations
- Tolloid-like 1
Tll-1
- Tolloid-like 2
Tll-2
- Turtle Tolloid
tTll
Footnotes
GenBank Accession numbers will be made available upon acceptance of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10(7):1001–10. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340(4):783–95. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bond JS, Beynon RJ. The astacin family of metalloendopeptidases. Protein Sci. 1995;4(7):1247–61. doi: 10.1002/pro.5560040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TG, Conway SJ, Scott IC, Labosky PA, Winnier G, Bundy J, Hogan BL, Greenspan DS. The mammalian Tolloid-like 1 gene, Tll1, is necessary for normal septation and positioning of the heart. Development. 1999;126(12):2631–42. doi: 10.1242/dev.126.12.2631. [DOI] [PubMed] [Google Scholar]
- Ferguson EL, Anderson KV. Localized enhancement and repression of the activity of the TGF-beta family member, decapentaplegic, is necessary for dorsal-ventral pattern formation in the Drosophila embryo. Development. 1992;114(3):583–97. doi: 10.1242/dev.114.3.583. [DOI] [PubMed] [Google Scholar]
- Gould E, Tanapat P. Stress and hippocampal neurogenesis. Biol Psychiatry. 1999;46(11):1472–9. doi: 10.1016/s0006-3223(99)00247-4. [DOI] [PubMed] [Google Scholar]
- Keifer J. In vitro classical conditioning of the turtle eyeblink reflex: approaching cellular mechanisms of acquisition. Cerebellum. 2003;2(1):55–61. doi: 10.1080/14734220310015610. [DOI] [PubMed] [Google Scholar]
- Keifer J, Clark TG. Abducens conditioning in in vitro turtle brain stem without cerebellum requires NMDA receptors and involves upregulation of GluR4-containing AMPA receptors. Exp Brain Res. 2003;151(3):405–10. doi: 10.1007/s00221-003-1494-5. [DOI] [PubMed] [Google Scholar]
- Keifer J, Sabirzhanov BE, Clark TG. Characterization of a novel reptilian Tolloid-like gene in brain of the pond turtle and expression after in vitro classical conditioning. Society for Neuroscience; Society for Neuroscience; Washington, D.C.: 2006. [Google Scholar]
- Liu Q-R, Hattar S, Endo S, MacPhee K, Zhang H, Cleary LJ, Byrne JH, Eskin A. A Developmental Gene (Tolloid/BMP-1) Is Regulated in Aplysia Neurons by Treatments that Induce Long-Term Sensitization. The Journal of Neuroscience. 1997;17(2):755–764. doi: 10.1523/JNEUROSCI.17-02-00755.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokin M, Keifer J. Targeting of GLUR4-containing AMPA receptors to synaptic sites during in vitro classical conditioning. Neuroscience. 2004;128(2):219–28. doi: 10.1016/j.neuroscience.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Mokin M, Lindahl JS, Keifer J. Immediate-early gene-encoded protein Arc is associated with synaptic delivery of GluR4-containing AMPA receptors during in vitro classical conditioning. J Neurophysiol. 2006;95(1):215–24. doi: 10.1152/jn.00737.2005. [DOI] [PubMed] [Google Scholar]
- Mullins MC. Holy Tolloido: Tolloid cleaves SOG/Chordin to free DPP/BMPs. Trends Genet. 1998;14(4):127–9. doi: 10.1016/s0168-9525(98)01431-0. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ. Evolutionary families of peptidases. Biochem J. 1993;290(Pt 1):205–18. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ. Evolutionary families of metallopeptidases. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- Scott IC, Blitz IL, Pappano WN, Imamura Y, Clark TG, Steiglitz BM, Thomas CL, Maas SA, Takahara K, Cho KWY, Greenspan DS. Mammalian BMP-1/Tolloid-Related Metalloproteinases, Including Novel Family Member Mammalian Tolloid-Like 2, Have Differential Enzymatic Activities and Distributions of Expression Relevant to Patterning and Skeletogenesis. Developmental Biology. 1999;213(2):283–300. doi: 10.1006/dbio.1999.9383. [DOI] [PubMed] [Google Scholar]
- Scott IC, Steiglitz BM, Clark TG, Pappano WN, Greenspan DS. Spatiotemporal expression patterns of mammalian chordin during postgastrulation embryogenesis and in postnatal brain. Developmental Dynamics. 2000;217:449–456. doi: 10.1002/(SICI)1097-0177(200004)217:4<449::AID-DVDY12>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Labosky PA, Furuta Y, Hargett L, Dunn R, Fogo AB, Takahara D, Peters DMP, Greenspan DS, Hogan BLM. Failure of ventral body wall closure in mouse embryos lacking a procollagen C-proteinase encoded by Bmp 1, a mammalian gene related to Drosophilia tolloid. Development. 1996;122(11):3587–95. doi: 10.1242/dev.122.11.3587. [DOI] [PubMed] [Google Scholar]
- Takahara K, Brevard R, Hoffman GG, Suzuki N, Greenspan DS. Characterization of a novel gene product (mammalian tolloid-like) with high sequence similarity to mammalian tolloid/bone morphogenetic protein-1. Genomics. 1996;34(2):157–65. doi: 10.1006/geno.1996.0260. [DOI] [PubMed] [Google Scholar]
- Takahara K, Lyons GE, Greenspan DS. Bone Morphogenetic Protein-1 and a Mammalian Tolloid Homologue (mTlD) Are Encoded by Alternative Spliced Transcripts Which Are Differentially Expressed in Some Tissues*. The Journal of Biological Chemistry. 1994;269(51):32572–32578. [PubMed] [Google Scholar]
- Tamura G, Olson D, Miron J, Clark TG. Tolloid-like 1 is negatively regulated by stress and glucocorticoids. Brain Res Mol Brain Res. 2005;142(2):81–90. doi: 10.1016/j.molbrainres.2005.09.016. [DOI] [PubMed] [Google Scholar]
- van Praag H, Christie BR, Sejnowski TJ, Gage FH. Running enhances neurogenesis, learning, and long-term potentiation in mice. PNAS. 1999;96(23):13427–13431. doi: 10.1073/pnas.96.23.13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Oelgeschlager M. Regulation of bone morphogenetic proteins in early embryonic development. Naturwissenschaften. 2004;91(11):519–34. doi: 10.1007/s00114-004-0575-z. [DOI] [PubMed] [Google Scholar]