SUMMARY
Little is known about how the distinct architectures of dendrites and axons are established. From a genetic screen, we isolated dendritic arbor reduction (dar) mutants with reduced dendritic arbors but normal axons of Drosophila neurons. We identified dar2, dar3, and dar6 genes as the homologs of Sec23, Sar1, and Rab1 of the secretory pathway. In both Drosophila and rodent neurons, defects in Sar1 expression preferentially affected dendritic growth, revealing evolutionarily conserved difference between dendritic and axonal development in the sensitivity to limiting membrane supply from the secretory pathway. Whereas limiting ER to Golgi transport resulted in decreased membrane supply from soma to dendrites, membrane supply to axons remained sustained. We also show that dendritic growth is contributed by Golgi outposts, which are found predominantly in dendrites. The distinct dependence between dendritic and axonal growth on the secretory pathway helps to establish different morphology of dendrites and axons.
INTRODUCTION
Dendrites and axons, two major compartments of neurons, exhibit a number of morphological distinctions. Little is known about how the structural differences between dendrites and axons are established. The growth of dendrites and axons likely involves coordinated cytoskeletal reorganization and membrane trafficking (Bradke and Dotti, 2000; Lecuit and Pilot, 2003). The cytoskeleton is organized differently in dendrites and axons (Baas et al., 1988). Furthermore, the cytoskeleton regulators RhoA (Lee et al., 2000) and MAP2 (Harada et al., 2002) preferentially control dendritic growth. It may also be important to regulate the membrane supply for dendrites and axons differently to establish their different morphology.
The secretory pathway, which includes the endoplasmic reticulum (ER), Golgi complex, and post-Golgi intermediates (van Vliet et al., 2003), is the major source of plasma membrane. Unique to neurons is the presence of an additional satellite secretory pathway, including ER and Golgi outposts, in dendrites (Aridor et al., 2004; Gardiol et al., 1999; Horton and Ehlers, 2003). Golgi outpost has so far not been found in axons (Horton and Ehlers, 2003), raising the question whether the polarized distribution of Golgi in neurons plays a role in the differential development of dendrites and axons. Recent findings suggest an active role of the polarized secretory trafficking in the formation of specific compartments of the dendritic arbors (Horton et al., 2005). Whether the secretory pathway also plays a role in distinguishing the growth of dendrites and axons is an important open question. Furthermore, whether dendritic Golgi outposts are required for dendritic growth and thus contribute to the generation of distinct dendrite and axon morphology is still unknown.
From a large scale genetic screen by taking advantage of a highly specific marker for the class IV dendritic arborization (da) neurons in Drosophila, we isolated 8 complementation groups of dendritic arbor reduction (dar) mutants, which share a common phenotype of reduced dendritic arbors but normal axonal growth. Strikingly, 3 of the 5 dar genes that we have so far identified molecularly encode critical regulators of the forward secretory pathway. Mosaic analysis revealed that these genes function cell-autonomously to regulate both dendritic growth and the secretory pathway. Similar functions of Sar1 were observed in cultured mammalian neurons, corroborating the notion that dendritic and axonal growth exhibit distinct sensitivity to changes in ER-to-Golgi transport. We further show that such distinct sensitivity involves preferential reduction of membrane transport from soma to dendrites upon global reduction in ER-to-Golgi transport, and requirement of Golgi outposts for dendritic growth.
RESULTS
Dendritic and axonal growth exhibit distinct sensitivity to the reduction of ER-to-Golgi transport in da neurons in vivo
We recently carried out a genetic screen to identify mutants with defects in dendritic and/or axonal development (Grueber et al., 2007) by taking advantage of the highly specific marker for the class IV da neurons in Drosophila peripheral nervous system (PNS), pickpocket-EGFP (ppk-EGFP) (Grueber et al., 2003). These neurons resemble mammalian neurons in morphological and cell biological features, such as multipolar morphology and the tapering of dendrites but not axons, making them a suitable model to study the differential development of dendrite and axon.
From 3299 3rd chromosomes carrying lethal mutations, we isolated 12 mutants that showed defects in dendritic but not axonal growth (Figure 1A). These 12 mutants fell into 8 complementation groups, which we named dendritic arbor reduction (dar) 1–8 (Table 1). Through genetic mapping and molecular characterization, we identified 3 dar genes, namely dar2, dar3, and dar6, as the Drosophila homologs of Sec23, Sar1, and Rab1, respectively (Figure S1 & S2 and Supplemental Experimental Procedures), all of which are critical regulators of ER-to-Golgi transport mediated by the COPII vesicles (Lee et al., 2004). Defects in the functions of Sar1 (Zaal et al., 1999), Rab1 (Wilson et al., 1994), or Sec23 (Morin-Ganet et al., 2000) result in dispersion of Golgi complexes. We focused on Sar1/Dar3 in subsequent studies, because Sar1 is the primary component that initiates the formation of COPII coated vesicles for forward trafficking from ER to Golgi.
Figure 1.
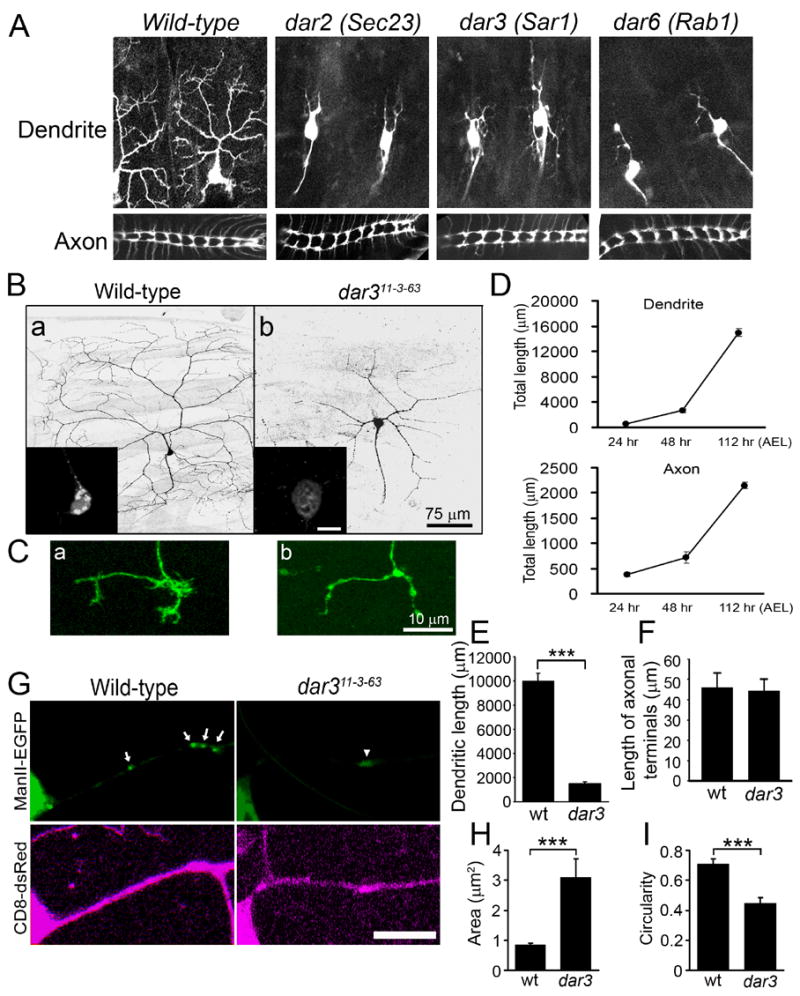
Dendritic and axonal growth exhibit distinct sensitivity to the reduction of ER-to-Golgi transport in da neurons in vivo. (A) Dendritic arbors (top panels) and axon terminal projections (bottom panels) of class IV da neurons in wild-type, dar2, 3, and 6 mutant embryos (stage 17). (B) Dendrite morphology of wild-type (a) and dar311-3-63 mutant (b) mosaic clones generated by Golgi-MARCM. Scale bar: 75 μm. The insets show the somatic Golgi structure labeled by ManII-EGFP. Scale bar: 10 μm. (C) Axonal terminals of wild-type (a) and dar311-3-63 mutant (b) ddaC neurons. Scale bar: 10 μm. (D) Time course of dendritic and axonal growth of class IV ddaC neurons (segment A5) during larval development. For larvae cultured at 25°C, 24 hr, 48 hr, and 112 hr after egg laying (AEL) correspond to early 1st, 2nd, and late 3rd instar, respectively. Error bars represent S.E.M. in this paper. N=3. (E) Quantitation of total dendritic length of wild-type (wt) and dar3 mutant ddaC neurons. The dendritic arbors of wild-type neurons that were quantified here did not include the fine braches close to the segment borders for the convenience of image collection. “*”, “**”, and “***” indicate p<0.05, 0.01, and 0.001, respectively, in this paper. t-test was used in this paper unless otherwise specified. (F) Quantitation of the length of axonal terminals excluding filopodium-like structures (p>0.05). (G) Discrete Golgi outpost structures are replaced by diffuse ManII-EGFP signal in the dendrites of dar311-3-63 mutant neurons. Arrows point to Golgi outposts in a wild-type neuron and the arrowhead points to diffuse ManII-EGFP signal in a dar3 mutant neuron. Scale bar: 10 μm. (H–I) Quantitation of the size (H) and circularity (4π X area/perimeter2) (I) of Golgi outposts in wild-type neurons and diffuse ManII-EGFP signal in dar3 mutant neurons.
Table 1.
dendritic arbor reduction mutants isolated from the screen.
| Mutants | Number of alleles | Cytological location/Gene | Cell-autonomy |
|---|---|---|---|
| dar1 | 2 | KLF transcription factor | Yes |
| dar2 | 1 | Sec23 | TBD |
| dar3 | 1 | Sar1 | Yes |
| dar4 | 1 | 93A1-93B3 | TBD |
| dar5 | 1 | 69C4-69D1 | No |
| dar6 | 2 | Rab1 | Yes |
| dar7 | 2 | 72D1-93D1 | TBD |
| dar8 | 2 | 63F6-7 or 63A6-9 | TBD |
TBD: to be determined.
We examined dar3 functions in da neurons by mosaic analysis with a repressible cell marker (MARCM) (Lee and Luo, 1999). We incorporated a Golgi marker, α-mannosidase II-EGFP (ManII-EGFP) (Velasco et al., 1993) into this analysis in addition to the membrane marker mouse CD8-dsRed (mCD8-dsRed) for marking neuron morphology. We refer to this assay as Golgi-MARCM.
The Golgi-MARCM analysis revealed that class IV da neurons with defective dar3 exhibited reduced dendritic arbors (Figure 1B), demonstrating that dar3 gene is cell-autonomously required for dendritic growth. The class IV neuron ddaC normally had a total dendrite length of 10,016 ± 633 μm (n=3; data represent mean ± standard error of means in this paper). The total dendritic length of dar3 mutant neurons was reduced to 1,488 ± 171 μm (n=3), only 14.9% of control (Figure 1E).
In contrast to the dramatic reduction in dendritic growth, the axonal growth appeared normal except for subtle changes in the terminals. As the animal increases from ~ 0.5 to ~ 4 mm in length from embryonic to 3rd instar larval stages, the axons of da neurons extend in proportion between the body wall and the ventral nerve cord (VNC), where short terminal projections form a simple arbor (Grueber et al., 2007). From early 1st instar stage (24 hr after egg laying, AEL) to late 3rd instar stage (112 hr AEL) the axons of the ddaC neurons in abdominal segment 5 (A5) grew from 384 ± 41 to 2,151 ± 64 μm (Figure 1D). During this period of extensive axonal growth, the axons of dar3-deficient ddaC neurons not only grew into the VNC but also formed projections that were comparable to those of wild-type neurons except that the short fibers which resemble filopodia were often missing (Figure 1C-b). In VNC, the ddaC (segments A3–A5) terminal projection contains one anterior, one contra-lateral, and, in some cases, one posterior branch (Figure 1C-a). Several (7.5 ± 0.7, n=4) short and fine fibers, resembling filopodia, originate from these terminal branches. The average total length of axonal terminals of dar3 mutant ddaC neurons (44.3 ± 5.5 μm, n=7), excluding the filopodium-like fibers, was comparable to that of wild-type ddaC neurons (45.8 ± 7.0 μm, n=4, p>0.05) (Figure 1F). The filopodium-like fibers, with a total length of 24.8 ± 3.5 μm (n=4), comprise only a small fraction of total axonal length (1.2%). In contrast to this minor reduction in filopodium-like structures at the axonal endings, the reduction of dendritic arbors in dar3 mutant neurons by 8,528 μm, which is 85.1% of the total dendritic length (Figure 1E), was much greater in extent.
Our Golgi-MARCM experiment also showed that discrete Golgi structures in the soma were replaced by diffuse distribution of the marker ManII-EGFP in dar3 mutant neurons (Figure 1B). Similarly, discrete, bright, and round/oval-shaped Golgi outposts in dendrites, labeled by ManII-EGFP, were replaced by weak and diffuse signal in neurons with defective dar3 (Figure 1G). The diffuse signal covered a greater area than Golgi outposts (3.09 ± 0.62 μm2 versus 0.84 ± 0.06 μm2 in wild-type neurons, P<0.001) (Figure 1H), and took on irregular shape with an average circularity of 0.45 ± 0.04, unlike Golgi outposts which normally have an average circularity of 0.71 ± 0.03 (P<0.001) (Figure 1I). Therefore, Sar1 is required for normal morphology of both somatic Golgi and dendritic Golgi outposts.
Dendritic and axonal growth exhibit distinct sensitivity to the reduction of ER-to-Golgi transport in cultured hippocampal neurons
In most neurons, the initiation of axonal growth precedes that of dendrites. One potential cause of the observed “dar” phenotype is that the maternally contributed Dar proteins in the embryos rescued defects in axonal growth but was insufficient to rescue dendritic growth. We eliminated maternal contribution by generating mosaic clones defective of dar3 or dar6 in germline cells but found that females with such mutant germline cells were sterile, thus precluding us from examining the dendrite phenotype of germline clones.
To test whether temporal differences between dendritic and axonal growth account for the “dar” phenotype, we need to alter dar gene function when both dendrites and axons actively grow. In the MARCM experiments described above, the da neurons were born during early embryogenesis and imaged several days later in 3rd instar larvae. During this period, both dendrites and axons grow extensively (Figure 1D). Despite the dramatic reduction in the dendrite length, these dar3-defective class IV da neurons had normal total axon length. These results suggest that dendritic and axonal growth indeed exhibit different dependence on the dar3 gene.
Another system in which the function of dar genes could be altered when both dendrites and axons actively grow is the hippocampal neuronal culture. Although the initiation of axonal growth is also earlier than dendrites, there is an extensive period in which both axons and dendrites actively grow after 2 days in vitro (DIV) (Dotti et al., 1988). In our system, dendritic length increased by 194.4 μm, and axonal length increased by 2939.9 μm from 2 (n=19) to 7 DIV (n=47) (Figure 2A). To interfere with ER-to-Golgi transport, we generated small interfering RNAs that specifically and effectively knocked down the expression of Sar1 (Sar1-siRNA) (Figure S3). After introduction into cultured neurons at 2 DIV, Sar1-siRNA reduced endogenous Sar1 protein level to 42% at 5 DIV compared to control neurons. Knockdown of Sar1 also caused dispersion of Golgi complex in cultured neurons (Figure 2B), consistent with findings from non-neuronal cells (Ward et al., 2001).
Figure 2.
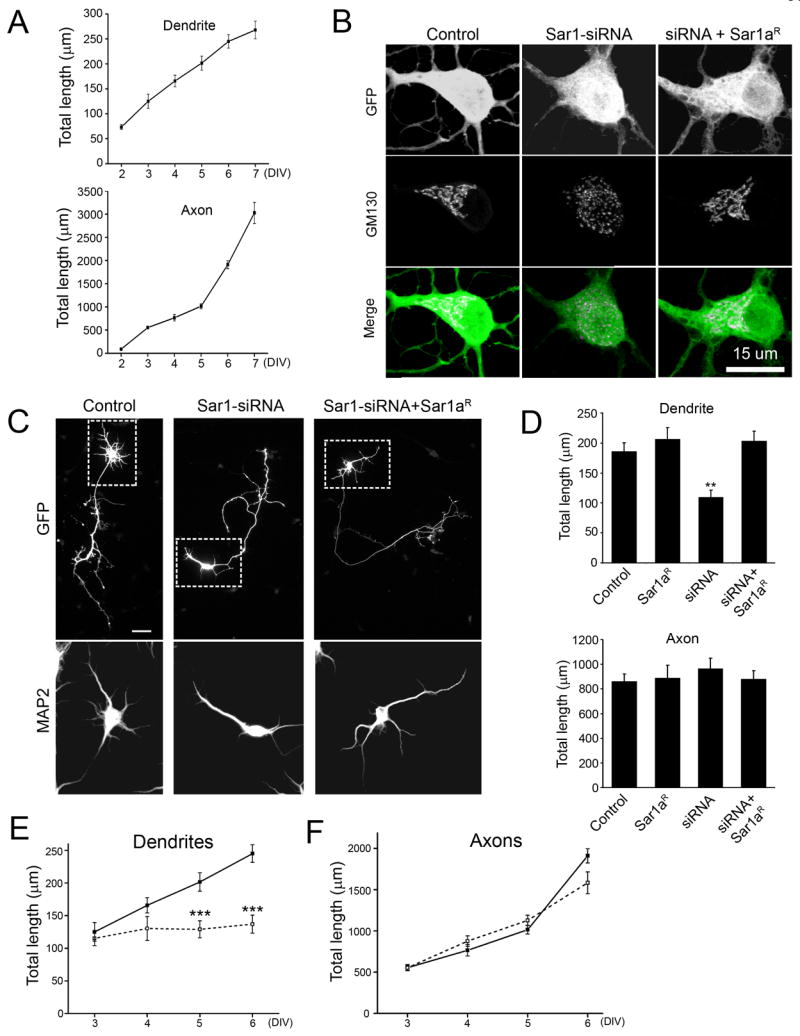
Dendritic growth is preferentially reduced in cultured hippocampal neurons with defective ER-to-Golgi transport. (A) Time course of dendritic and axonal growth in cultured hippocampal neurons. Neurons transfected with a plasmid encoding EGFP were stained with anti-MAP2 and anti-GFP antibodies, together with the nuclear dye DAPI. The neurites positive for MAP2 were identified as dendrites. The neurites negative for MAP2 were identified as axons. Only neurons with normal nuclear morphology as revealed by DAPI-stain were included for quantitation to eliminate dying neurons. (B) Sar1-siRNAs dispersed Golgi complex in cultured hippocampal neurons and a siRNA-resistant construct of Sar1a (Sar1aR) rescued Golgi defects. The Golgi complex was labeled with anti-GM130 antibody. (C) Neurons transfected with Sar1-siRNA exhibit dramatically reduced dendritic growth and a siRNA-resistant Sar1a (Sar1aR) construct restored dendritic growth. Scale bar: 50 μm. The somatodendritic regions are boxed with dashed lines. The images of MAP2 staining for the boxed regions are shown in the bottom panel. (D) Quantification of dendritic and axonal growth in cultured hippocampal neurons. N= 37, 9, 15, and 24 for control, Sar1aR, siRNA, and siRNA+ Sar1aR, respectively. ANOVA test. The experiments were carried out in a double-blind fashion. (E–F) Time course of the effect of Sar1-siRNA on dendritic and axonal growth. The mean total dendritic (E) and axonal (F) length of control (solid line) and Sar1-siRNA-transfected (dashed line) neurons was shown.
Reducing Sar1 protein level in hippocampal neurons dramatically decreased the total length of dendrites but not axons as revealed by double-blind analysis (Figure 2C and D). At 5 DIV, total dendritic length of control and Sar1-siRNA-transfected neurons were 185.6 ± 15.0 μm and 109.7 ± 11.7 μm, respectively (P<0.01, ANOVA), while total axonal length were 861.3 ± 60.1 μm (n=37) and 964.7 ± 86.0 μm (n=15) (p>0.05, ANOVA), respectively. A siRNA-resistant Sar1a construct (Sar1aR) rescued the dendrite and Golgi defects (Figure 2B–D & S3) (dendrite length=203.1 ± 16.8 μm, p>0.05; axon length=880.8 ± 67.9 μm, P>0.05, ANOVA), suggesting the siRNA knockdown was specific. Comparison of dendrite and axon length between control and Sar1-siRNA-expressing neurons at 3, 4, 5, and 6 DIV revealed that dendrites ceased to extend following knockdown of Sar1 (Figure 2E) while axons continued to grow normally (Figure 2F). Thus, reducing Sar1 expression in hippocampal neurons significantly reduced dendrite length but not axon length, revealing different dependence of dendritic and axonal growth on the secretory pathway in mammalian neurons as in Drosophila neurons.
Supply of membrane labeled by mCD8-GFP was adjusted unequally between dendrites and axons in response to perturbation of secretory pathway function
To test how the membrane supply to dendrites and axons may be adjusted upon reducing the activity of secretory pathway, we employed fluorescence recovery after photo-bleaching (FRAP) analysis in rat hippocampal neurons and examined the effects of Sar1-siRNA. FRAP monitors the traffic of molecules into a selected area that has previously been photo-bleached (Lippincott-Schwartz et al., 2001). To examine membrane supply from soma to dendrites and axons, including both intracellular vesicle transport and lateral diffusion of plasma membrane, we marked membrane with the fluorescent transmembrane protein mCD8-EGFP. Photo-bleaching of the EGFP signal was carried out for the entire dendritic and axonal arbors but not the soma. The fluorescence recovery in dendrites and axons, due to supply from the soma, was monitored immediately following photo-bleaching over a 15-min period. We sampled fluorescence recovery at proximal dendrites and axons (5 μm away from the soma) as an indicator of the amount of membrane passing through this segment as well as the limited amount of membrane being added to this segment (Figure 3A). Control neurons exhibited comparable rate and extent of fluorescence recovery in dendrites and axons (Figure 3B). However, Sar1-siRNA significantly reduced the delivery of fluorescent membrane proteins from soma to dendrites, without affecting the supply to axons (Figure 3B), as evident from quantitation of the maximal percentage of recovery (control dendrites: 27.0 ± 1.7; Sar1-siRNA dendrites: 18.6 ± 2.0; P<0.01, t-test; control axons: 26.5 ± 2.7; Sar1-siRNA axons: 25.7 ± 4.6; P>0.05) and the time constant (control dendrites: 129.7 ± 13.4 sec; Sar1-siRNA dendrites: 185.6 ± 25.5 sec; P<0.05, t-test; control axons: 118.6 ± 22.1 sec; Sar1-siRNA axons: 113.1 ± 26.6 sec; P>0.05) (Figure 3C). These results suggest that the membrane supply from soma to dendrites and axons, at least that labeled by mCD8-GFP, is regulated in an unequal way upon reduction in secretory pathway activity.
Figure 3.
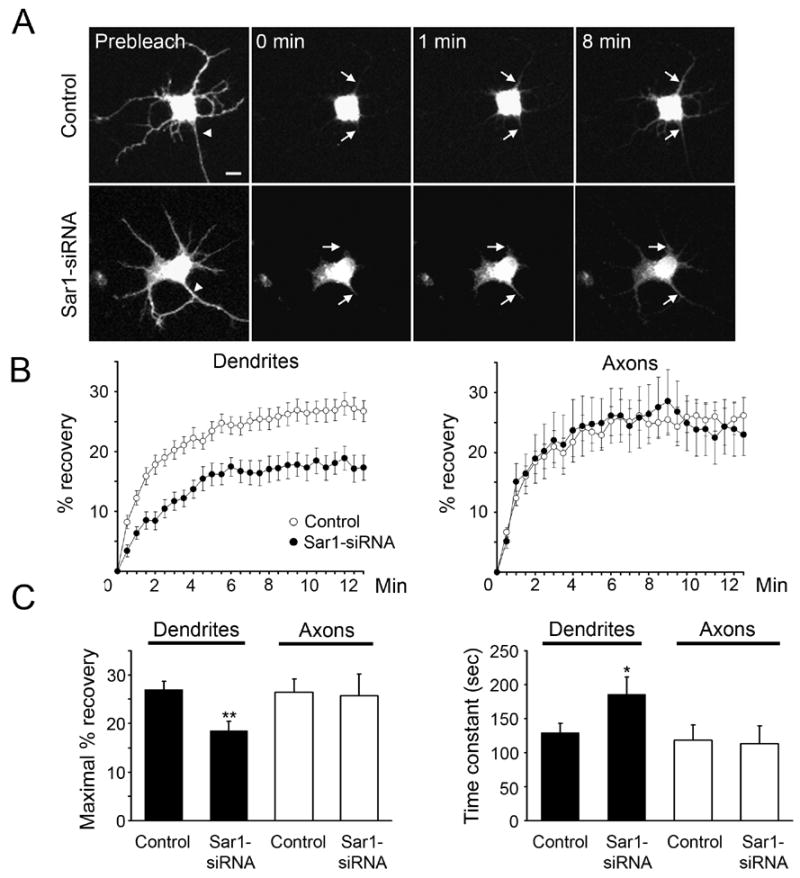
Limiting ER-to-Golgi transport leads to preferential reduction of dendritic membrane supply. (A) Representative pictures of membrane-bound EGFP (mCD8-EGFP) moving from soma to dendrites and axons during the FRAP experiments. Zero min is defined as immediately after bleaching. Arrowheads point to the axon. Arrows point to areas that are 5 μm away from soma in representative dendrites and axons. Scale bar: 10 μm. (B) Time course of recovery of mCD8-EGFP signals in photo-bleached dendrites and axons. Open circles: control neurons (n=26); filled circles: Sar1-siRNA-transfected neurons (n=12). (C) Maximal fluorescence recovery is reduced, while time constant is increased, in the dendrites but not axons of the siRNA-transfected neurons.
Directional movements of dendritic Golgi outposts correlate with the extension and retraction of dendritic branches
The secretory pathway in neurons contains not only ER and Golgi in the soma, but also a satellite secretory pathway in dendrites (Gardiol et al., 1999; Horton and Ehlers, 2003; Pierce et al., 2001). Given the enrichment of Golgi outposts in dendrites over axons in mammalian neurons (Horton and Ehlers, 2003), we explored the possibility that Golgi outposts contribute to dendritic growth. In class IV da neurons, Golgi outposts appeared as punctate structures in both major and fine terminal dendritic branches but rarely in the axons (Figure 4A & C). They were often enriched at dendritic branching points, While the primary dendrites contained 10.34 ± 1.34 (n=13) Golgi outposts per 100 μm, the proximal 100 μm of the axon contained only 1.00 ± 0.32 (n=5) (Figure 4B).
Figure 4.
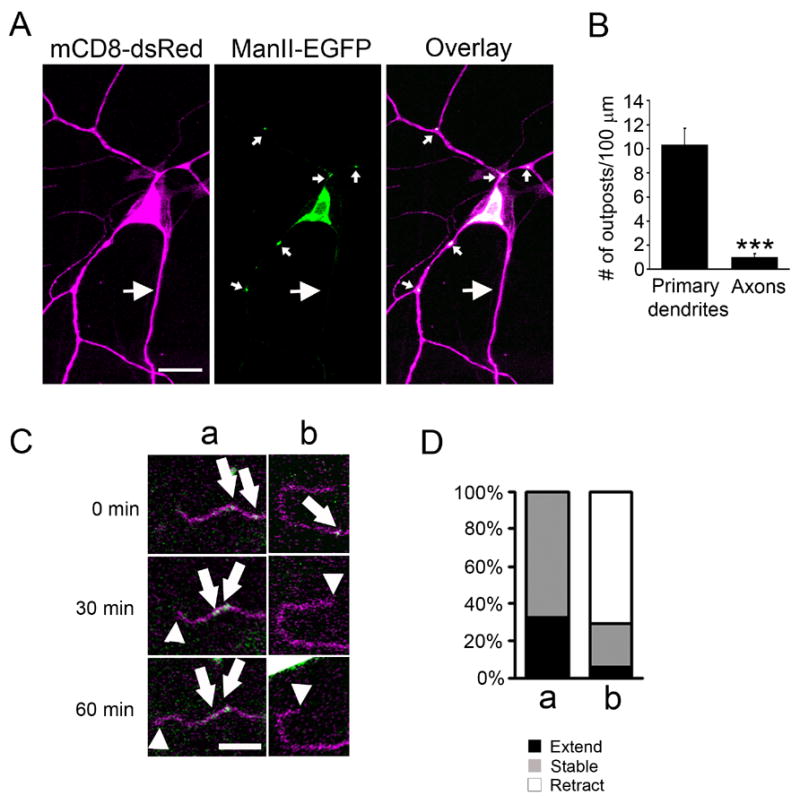
Golgi outpost dynamics correlates with dendritic branch dynamics. (A) A class IV da neuron expressing mCD8-dsRed (magenta) and ManII-EGFP (green). Golgi complex are present in the soma and dendrites, but rarely seen in the axon. Small arrows point to dendritic Golgi outposts and large arrows point to the axon. Scale bar: 15 μm. (B) Quantitation of the number of outposts in primary dendrites and the proximal 100 μm of axon. Primary dendrites were defined as those that originated from the soma and ended at the first branch point. (C) Time-lapse imaging of the movement of Golgi outposts and dendritic dynamics. The panels show that Golgi outposts moved distally prior to branch extension (a) and disappeared prior to branch retraction (b). Arrows point to the Golgi outposts that appeared, disappeared or moved. Arrow heads point to the tip of the branch. Scale bar: 5 μm. (D) Quantitation of the correlation between Golgi outpost dynamics and branch dynamics. a, Golgi outposts move distally or appear de novo in a branch; b, Golgi outposts move proximally or disappear in a branch.
To characterize the involvement of Golgi outposts in dendritic growth, we monitored the behavior of Golgi outposts during branch extension and retraction by time-lapse imaging in live Drosophila larvae. We found a strong correlation between Golgi outpost dynamics and dendritic branch dynamics. Specifically, a branch either extended or remained stable after the Golgi outposts moved distally or appeared de novo within the branch (Figure 4C& D) (extension: 32.4%; stable: 67.6%; retraction: 0%; n = 34), while the majority of the branches retracted after the Golgi outpost moved proximally or disappeared (Figure 4C & D) (extension: 5.9%; stable: 23.5%; retraction: 70.6%; n=17). An interesting implication of this finding is that dendritic Golgi outposts could contribute to the supply of the membrane needed for branch growth in their vicinity. Outposts move distally or proximally prior to the extension or retraction of the dendritic branches, serving as a mobile station for membrane traffic.
Dendritic Golgi outposts are important for the dynamics of dendritic branches
To test whether the function of Golgi outposts is required for dendritic growth, we attempted to damage Golgi outposts with laser. Terminal branches of class IV da neurons are highly dynamic. In 3rd instar larvae, 20.8 ± 5.9% branches showed extension (>0.5 μm) and 41.7 ± 7.1% branches showed retraction (>0.5 μm) over a period of just 5 minutes (n=48) (Figure 5B & C). After Golgi outposts labeled by β-1,4-galactosyltransferase:EYFP (GalT-EYFP) were focally illuminated by intense laser, most of the branches did not extend or retract (percentage of branches that extended = 5.0 ± 4.9%, percentage of branches that retracted = 5.0 ± 4.9%, n=20). To confirm that the reduction of branch dynamics resulted from damage to Golgi outposts, we illuminated dendritic regions lacking a Golgi outpost (extension: 22.6 ± 7.5%; retraction: 38.7 ± 8.8%; n=31), regions labeled by actin-GFP (extension: 30.0 ± 10.3%; retraction: 25.0 ± 9.7%; n=20), or mito-GFP (a mitochondria marker) (Cox and Spradling, 2003) (extension: 23.8 ± 9.3%; retraction: 23.8 ± 9.3%; n=21),with the same laser energy used in damaging Golgi outposts (Figure 5A). Neither the extension nor the retraction of dendritic branches showed significant change after any of these control illuminations compared to dendrites without laser illumination (p>0.05) (Figure 5B & C). Despite that the nature and extent of laser damages to Golgi outposts remain unclear, Golgi outposts appeared to be the most sensitive locations for affecting dendritic branch dynamics among all the targets tested. We thus conclude that dendritic Golgi outposts are important for the extension and retraction of dendritic branches.
Figure 5.
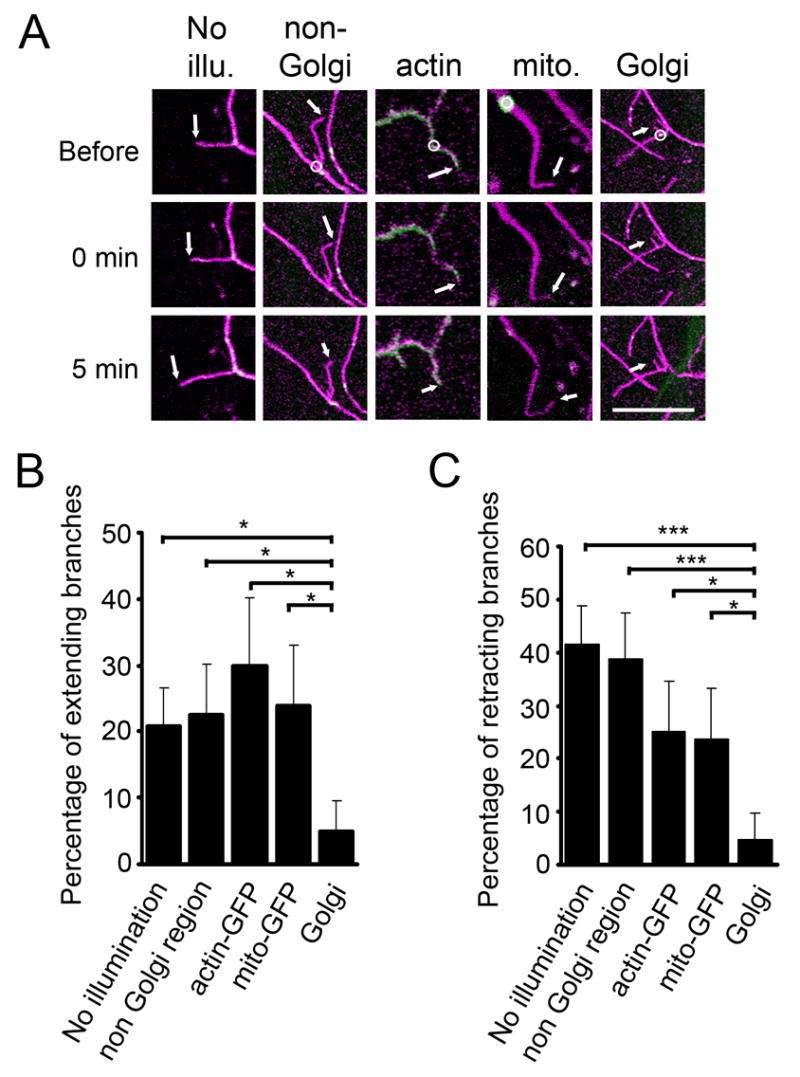
Laser damaging of dendritic Golgi outposts reduces the extension and retraction of dendritic branches. (A) Examples of dendritic branches without laser illumination (No illu.), after focal intense laser illumination on regions lacking Golgi outposts (non-Golgi), on regions with actin-GFP (actin), on mitochondria (mito.), and on Golgi outposts (Golgi). Magenta: mCD8-dsRed. Green: actin-GFP in “actin”, mito-GFP in “mito.”, GalT-EYFP in the other 3 panels. Upper, middle, and lower panels represent images before, immediately after, and 5 minutes after laser damaging, respectively. Circles indicate the regions of laser illumination. Arrows point to the tips of the branches being tested. Scale bar: 10 μm. (B–C) Percentage of branches that extended (B) or retracted (C) after each manipulation.
Redistribution of dendritic Golgi outposts correlates with morphological changes of dendritic arbors
To further test whether Golgi outposts are critical for dendritic growth, we examined the effect of redistributing Golgi outposts. The positioning of Golgi complex involves several molecular motors, among which dynein has the major role (Allan et al., 2002). The cargo specificity of dynein relies on the dynactin complex (Schroer, 2004). In Drosophila, Lava lamp (Lva) mediates the interactions between Golgi and the dynein/dynactin complex (Papoulas et al., 2005).
We found that Lva protein was expressed in da neurons and was located juxtaposed to ManII-EGFP, which labels the medial/trans-Golgi (Figure S4). We reasoned that blocking the function of Lva could impair the transport and thus change the distribution of Golgi outposts in da neurons. We took two different approaches to test this. First, we expressed a dominant negative form of Lva (LvaDN) in class IV da neurons to disrupt Lva-dynactin interaction. LvaDN is a coiled-coil portion of Lva that binds the dynactin complex and has been shown to interfere with dynein-based movement of Golgi during cellularization (Papoulas et al., 2005). Second, we knocked down Lva expression using inducible RNAi. To achieve RNAi-mediated knockdown, we generated two transgenic lines that expressed inverted repeats, each targeting a distinct part of the lva mRNA (UAS-Lva-RNAi1 and UAS-Lva-RNAi2, see Supplemental Experimental Procedures), and tested an additional line from the Vienna Drosophila RNAi center (VDRC). All three lines showed similar phenotypes.
When expressed in class IV da neurons, both LvaDN and Lva-RNAi1 altered the distribution of Golgi outposts. In the distal part of dendritic arbors, both LvaDN and Lva-RNAi1 reduced the size of Golgi outposts (control: 1.66 ± 0.24 μm2 per 100 μm of dendrites, n=6; LvaDN: 0.49 ± 0.21 μm2, n=5; Lva-RNAi1: 0.59 ± 0.18 μm2, n=8; p<0.01) (Figure 6A & B). In proximal dendrites, LvaDN caused an increase of the size of Golgi outposts whereas Lva-RNAi1 caused a reduction (control: 5.46 ± 1.27 μm2 per 100 μm of dendrites, n=11; LvaDN: 10.83 ± 1.36 μm2, n=20; Lva-RNAi1: 2.47 ± 0.67 μm2, n=13; p<0.05) (Figure 6A & C). No change in the number and size of somatic Golgi was observed in either LvaDN- or Lva-RNAi1-expressing neurons (Figure 6D & S5).
Figure 6.
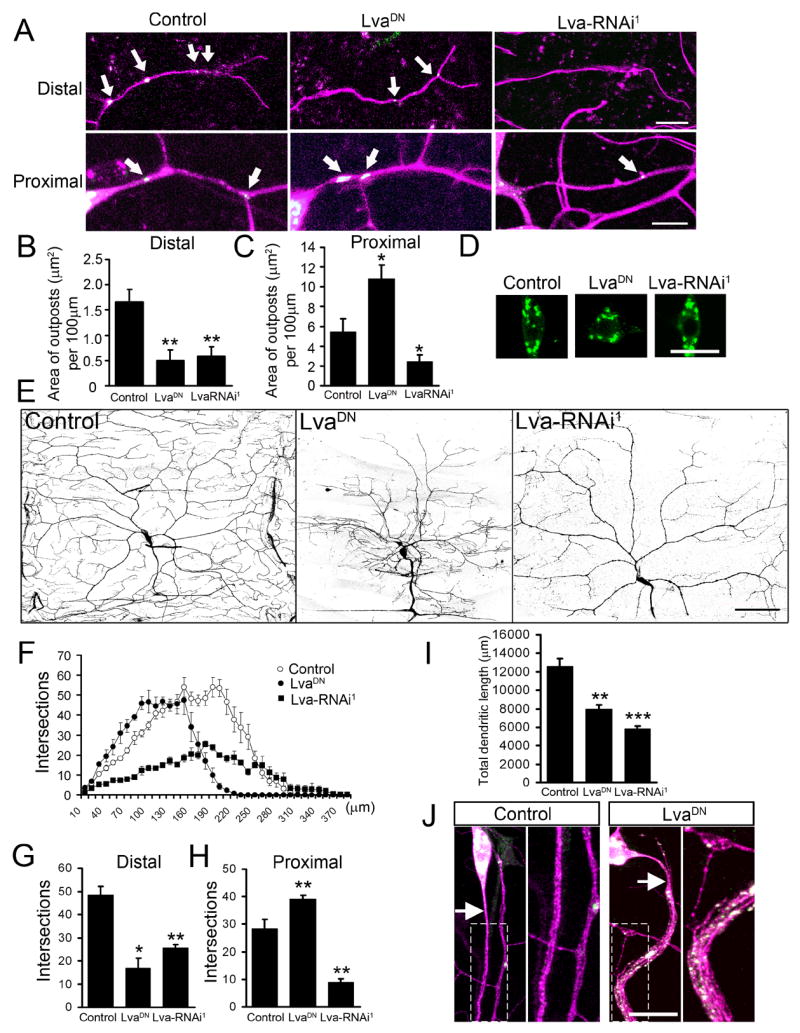
Redistribution of Golgi outposts by LvaDN and Lva-RNAi correlates with morphological changes of dendritic arbors. Magenta: mCD8-dsRed. Green: GalT-EYFP. (A) Representative pictures of Golgi outposts in distal and proximal dendrites in control, LvaDN-, and Lva-RNAi1-expressing ddaC neurons. Expression of LvaDN was driven by Gal44-77. Expression of Lva-RNAi was driven by Gal4109-2-80 to achieve earlier and stronger knockdown effect. Arrows point to Golgi outposts. Scale bars: 10 μm. (B–C) Quantification of the total size of Golgi outposts in distal dendrites (100 μm in length from the tip of the dorsal-most branch toward the soma) and proximal dendrites (30 μm from the soma). (D) Golgi structure in the soma. Scale bar: 15 μm. (E) Dendrite morphology of control, LvaDN-, and Lva-RNAi1-expressing ddaC neurons. Dendrites of Lva-RNAi1-expressing neurons were labeled by ppk-EGFP. Scale bar: 75 μm. (F) Sholl analysis histogram. (G) Number of intersections between distal dendritic branches and circles with 180 μm radius. (H) Number of intersections between proximal dendritic branches and circles with 80 μm radius. (I) Quantitation of total dendritic length. (J) Golgi outposts and exuberant branches appear in the proximal axons of LvaDN-expressing neurons. Magnified view of the boxed areas is shown in the right panels. The arrows point to the axons. Scale bar: 15 μm.
Parallel to the redistribution of Golgi outposts, there were changes in dendritic branches in the LvaDN- and Lva-RNAi1-expressing neurons (Figure 6E). In both LvaDN- and Lva-RNAi1-expressing neurons, the number of dendritic branches in distal dendrites was dramatically reduced, while that in proximal dendrites was increased in LvaDN-expressing neurons and decreased in Lva-RNAi1-expressing neurons. Alterations in dendritic growth were quantified by the Sholl analysis (Sholl, 1953) (Figure 6F), which determines the number of intersections between dendritic branches and circles centered at the soma. The number of intersections between distal dendrites and a 180 μm-radius circle was 48.5 ± 3.7 in control (n=4), 17.0 ± 4.3 in LvaDN (n=4) (p < 0.05), and 25.5 ± 1.4 in Lva-RNAi1 (n=4) (p < 0.01) (Figure 6G). The number of intersections between proximal dendrites and an 80 μm-radius circle was 28.5 ± 3.3 in control (n=4), 39.0 ± 1.4 in LvaDN (n=4) (p < 0.01), and 9.0 ± 1.2 in Lva-RNAi1 (n=4) (p < 0.01) (Figure 6H). As a result, total dendritic length was reduced in both LvaDN- and Lva-RNAi-expressing neurons (control: 12,600 ± 810 μm; LvaDN: 7,964 ± 452 μm, p<0.01; Lva-RNAi: 5,828 ± 351 μm, p<0.001) (Figure 6I). Similar results were obtained with UAS-Lva-RNAi2, which targets a distinct region of lva mRNA, and the VDRC line, confirming the specificity of the RNAi effects (data not shown). Thus, the effects of LvaDN and Lva-RNAi on dendritic branching (Figure 6G & H) correlated very well with the effects on Golgi outpost redistribution (Figure 6B & C).
Interestingly, numerous Golgi outposts were also found in proximal axons of neurons expressing LvaDN (Figure 6J). Parallel to this, exuberant branching was observed in the proximal segment of the axons but not in the middle and distal segments. These branches are likely axons rather than ectopic dendrites because the dendritic marker, Nod: βGalactosidase (Nod: βGal) (Clark et al., 1997), was absent from these branches (Figure S6). We also examined the distal axonal projection pattern and found that the axons in LvaDN expressing neurons reached the VNC and formed normal projection patterns (data not shown).
Taken together with the correlation between dendritic growth dynamics and Golgi outpost movement and the impaired dendritic branch dynamics caused by laser-damaging of Golgi outposts, these results underscore the importance of these components of dendritic satellite secretory pathway in dendritic growth.
DISCUSSION
In the present study, we demonstrated that growing dendrites and axons display different sensitivity to changes in the activity of the secretory pathway. Our findings add to the accumulating evidence that the secretory pathway is involved in cell polarity and provide one of the first evidence of the importance of the satellite secretory pathway in dendrite development.
Roles of the secretory pathway in cell polarity
The secretory pathway is important for cell polarity. For example, during the cellularization of Drosophila embryos, membrane growth is tightly controlled in a polarized fashion (Lecuit and Wieschaus, 2000). New membrane produced through the secretory pathway is predominantly added to the apical side early in polarization and to the lateral side at later stages. Other examples of targeted membrane transport during cell polarization include the apical-basolateral polarization of Madin-Darby canine kidney epithelial cells (Simons and Fuller, 1985), cell cycle of the budding yeast (Finger et al., 1998), and cell migration (Kupfer et al., 1982). Thus, membrane trafficking is tightly controlled in order to supply membrane to specific subcellular compartments of polarized cells.
The establishment of dendritic and axonal arbors is a major compartmentalization process of neurons. Before dendrites and axons are formed, selective delivery of post-Golgi vesicles to one neurite precedes the specification of that neurite as the axon (Bradke and Dotti, 1997). After axon specification, large amount of plasma membrane is added to these two compartments with distinct architecture. Whether the secretory pathway is involved in the differential growth of dendrites and axons is an important but poorly understood question. Horton et al. (2005) found that blockade of post-Golgi trafficking by expressing a dominant-negative protein kinase D1 (PKD-KD) causes a preferential reduction in dendritic growth. Although this result may appear similar to our results from manipulation of Sar1 function, there is in fact an important difference. PKD is required for cargo trafficking to the basolateral but not the apical membrane in epithelial cells (Yeaman et al., 2004). Since the targeting of dendritic proteins shares some of the mechanisms of the basolateral protein transport in epithelial cells (Dotti and Simons, 1990), PKD-KD may only affect dendrite-specific cargo transport. In this study, we assessed how membrane resource is allocated between growing dendrites and axons when global membrane production is limited. It is possible that the efficiency of dendrite-and axon-specific transport, including those mediated by PKD, changes differently in response to global restriction of membrane resource.
Distinct membrane dynamics in growing dendrites and axons
The plasma membrane of cells can be inserted via exocytosis and internalized through endocytosis. The expansion of plasma membrane in growing dendrites and axons is achieved through the interplay between these two antagonistic processes. Before dendrites are formed in cultured hippocampal neurons, membrane is selectively added to the growth cone of growing axons as well as minor neurites that will later become dendrites (Craig et al., 1995). In more mature neurons, there is insertion of new membrane to the axonal growth cone but not to the dendritic growth cone (Craig et al., 1995). This raises the possibility that plasma membrane is added to either multiple sites along the dendritic surface or throughout the dendritic surface. It is conceivable that dendritic Golgi outposts play a role in such membrane addition given their requirement for dendritic growth as shown in this study.
It has been reported that plasma membrane of cultured hippocampal neurons is internalized throughout the dendrites but only in the presynaptic terminals in axons (Parton et al., 1992). Parton et al. also found that essentially all internalized structures, from both dendritic and axonal surface, move in the retrograde direction to the soma. We compared the rate of membrane removal from dendritic and axonal surfaces and found that, consistent with Parton et al. (1992), endocytosis is more prominent in dendrites than in axons (Figure S8). The plasma membrane of dendrites was endocytosed 8.27 ± 1.79 (n=3) times faster than that of axons at 3 DIV. Thus, the demand for membrane supply to dendrites is likely greater than what appears based on the length of dendrites.
The difference in the ways that membrane is supplied to growing dendrites and axons is poorly understood. Notwithstanding much progress made on dendritic traffic of the temperature sensitive mutant of the vesicular stomatitis virus glycoprotein (VSVG) fused to GFP (Horton and Ehlers, 2003; Horton et al., 2005), the preferential targeting of VSVG to dendrites (Dotti and Simons, 1990) precludes its application for comparing dendritic and axonal membrane dynamics. In this work, we compared the membrane supply from soma to dendrites and axons using the FRAP analysis. We sampled fluorescence recovery in the dendritic and axonal segment 5 μm away from the soma as an indicator of the amount of the membrane passing though this segment. The sampling approach was taken because even with moderate magnification (20X) only a fraction of the arbors can be included in the imaging field, precluding the monitoring of fluorescence recovery along the entire dendritic and axonal arbor. This FRAP analysis revealed that, when the secretory pathway function is limiting, membrane supply from soma to growing dendrites is preferentially affected.
Notably, the secretory pathway with reduced activity could still provide sufficient membrane to support axonal growth at least during the period described in this study. It is possible that there are mechanisms involving post-Golgi carriers to allocate different amount of membrane to dendrites and axons. Alternatively, endocytosed dendritic membrane can also support axonal growth via the transcytosis pathway (Bartheld, 2004). Lastly, trafficking that bypasses COPII-based ER to Golgi transport (Hasdemir et al., 2005), although not widely observed, might also contribute to axonal growth.
Function of dendritic Golgi outposts in dendritic growth
The finding that Golgi outposts are present in dendrites has generated great interests in the function of these structures. Two main functions have been proposed for Golgi outposts. One hypothesis is that they are involved in local protein translation (Steward and Schuman, 2003). This is supported by the presence of protein synthesis machinery in the dendrites (Steward and Levy, 1982; Steward and Reeves, 1988) and findings of local translation of membrane proteins in dendrites (Eberwine et al., 2001; Raab-Graham et al., 2006). Recently, Horton et al. (2005) suggest that Golgi outposts, together with somatic Golgi complex, participate in forming the apical dendrites of pyramidal neurons.
In this study, we monitored the dynamics of Golgi outposts in intact neurons of live Drosophila larvae and observed that the direction of outpost movement correlates with dendrite branch dynamics. This suggests that Golgi outposts are probably involved in both the extension and retraction of a dendritic branch. Indeed, laser damaging of the outposts in a branch reduced branch dynamics, making the dendrite excessively stable, which will lead to retarded growth during the expansion of dendritic arbors. It should be noted that, in our laser damaging experiments, the nature and extent of damages to Golgi outposts are unclear. It is indeed technically challenging to manipulate organelle function in a small, localized subcellular region. We consider the laser damaging experiment one of the first attempts of such manipulations. Although we can conclude that Golgi outposts are the most sensitive locations for affecting dendritic branch dynamics among the various target locations, alternative approaches are needed to test the function of Golgi outposts. In the present study, we complemented the laser damaging experiment with genetic manipulations to redistribute Golgi outposts. We found that dendritic branching patterns changed with the redistribution of Golgi outposts by LvaDN and Lva-RNAi. High level of LvaDN expression led to the strongest defect in Golgi outpost distribution and dendritic branching pattern (Figure S7). Although it is difficult to rule out the contribution of non-specific effects of LvaDN, there is very good correlation between the distribution of Golgi outposts and that of dendritic branches in neurons with varying extent of disruption of Lva function. These findings underscore the importance of Golgi outposts in dendritic growth.
It remains unclear how Golgi outposts are produced, transported to dendrites, and largely excluded from axons. Our results on Lva suggest that they are probably produced in the soma and transported to dendrites. The presence of Golgi outposts in axons of LvaDN-expressing neurons suggest that they might be actively transported out of axons under normal condition. The absence of Golgi outposts and exuberant branches in the proximal axons of neurons expressing Lva-RNAi is likely due to partial interference of Lva function as similar phenotype was seen in neurons expressing low level of LvaDN (Figure S7). The increase in the size of Golgi outposts and dendritic branches in neurons expressing high level of LvaDN possibly reflect the requirement of Lva-dynactin interaction for the budding of Golgi outposts, a phenomenon that we have observed (data not shown).
Although it is difficult to rule out the presence of outposts that are below the detection sensitivity in this study, axonal Golgi outposts, if any, must be dramatically smaller and/or fewer than dendritic outposts. Therefore, the requirement of Golgi outposts for dendritic growth provides a mechanism to differentially control dendrite and axon growth.
The secretory pathway is possibly under regulation to control dendritic growth
The distinct dependence of dendrites and axons on the secretory pathway, as well as the involvement of Golgi outposts for local dendritic dynamics, raise the possibility that the secretory pathway may be regulated to influence the elaboration of dendrites. Such regulation might involve both genetic programs for specifying intrinsic differences in dendritic patterning of different neurons and activity-dependent modifications of dendritic arbors.
Several molecules are known to be specifically involved in dendritic but not axonal growth. Some of these molecules function in a neural activity-dependent fashion, such as the calcium/calmodulin-dependent protein kinase IIα (CaMKIIα), the transcription factor NeuroD (Gaudilliere et al., 2004), and the Ca2+-induced transcriptional activator CREST (Aizawa et al., 2004), while others are neural activity-independent such as bone morphogenetic protein 7 (BMP-7) (Lein et al., 1995) and Dasm1 (Shi et al., 2004). It will be important to find out whether the secretory pathway contributes to their regulation of dendritic growth.
In summary, we have demonstrated that dendritic and axonal growth exhibit different sensitivity to changes in membrane supply from the secretory pathway. Our findings raise a number of questions regarding membrane trafficking in dendrite and axon development. Answers to these questions will provide cell biological basis for understanding how the tremendous diversity of neuron morphology is achieved during development and how the changes in morphology happen in pathological conditions.
EXPERIMENTAL PROCEDURES
Genetic screen for dar mutants
The procedure for establishing mutagenized Drosophila lines and imaging of fly embryos for da neuron dendrites and axons are described in Grueber et al., 2007 in detail.
Golgi-MARCM
To image neuron morphology and Golgi structure in the same neuron using MARCM analysis, we crossed hs-flippase; UAS-mCD8-dsRed; FRT82B tubulin-Gal80 flies with either GAL4109-2-80, UAS-ManII-EGFP; FRT82B flies to generate control clones or GAL4109-2-80, UAS-ManII-EGFP; FRT82B dar311-3-63/TM6B, Tb to generate mutant clones. Mosaic clones were generated as described previously (Ye et al., 2004) and examined either by live imaging or staining with rat anti-mCD8 (for neuron morphology), chicken anti-GFP (for Golgi complex), and goat anti-HRP crosslinked to Cy5 (for the morphology of the entire PNS).
Fluorescence recovery after photo-bleaching (FRAP)
Cultured neurons were transfected with pCDNA3 or Sar1-siRNAs together with mCD8-EGFP and mCherry (Shaner et al., 2004) at 2 DIV and used for FRAP experiments at 5 or 6 DIV. The neurons were maintained in HEPES-based artificial cerebrospinal fluid (ACSF) at 35 °C during experiment. FRAP was carried out with a Zeiss LSM510 confocal system with a 20X lens. EGFP signal in dendrites and axons, but not the soma, was photo-bleached by applying a 488 nm laser (66% of full power; 30 mW) for 6.4 μsec per pixel with 150–180 iterations. Immediately after photo-bleaching, images of EGFP-fluorescence were acquired every 30 sec. Intensity of EGFP signal in dendrites and axons was measured at a point 5 μm away from the soma with ImageJ software and the background was subtracted. Only neurites with more than 80% bleaching were used for quantitation. Percent recovery at time point t (Rt) was calculated as (It−I0) x100/(Ii−I0) where It, I0, and Ii are the fluorescence intensity at time point t, immediately after photobleaching, and before bleaching, respectively. The recovery time course was fit with the following equation by the OriginPro software (OriginLab Co., Northampton, MA): Rt=R∞ x (1−e−t/τ), where Rt is the percentage recovery, R∞ is the maximal recovery, t is time in seconds, and τ is the time constant.
Live imaging of dendritic Golgi outposts
To study the correlation between Golgi outpost movement and dendritic branch dynamics, 3rd instar larvae were mounted in halocarbon oil and imaged with a Zeiss LSM510 confocal system. Images were taken every 30 minutes for 2–6 hours. The larvae were released to a grape juice agar plate between imaging sessions. We followed the movement of Golgi outposts labeled by GalT-EYFP between the tip of a terminal branch and the branch point. When no outpost is present in the branch, we followed the outpost closest to the branch point. The movement of Golgi outposts was counted if one of the outposts moved in or away from a branch, regardless of the presence of remaining stable outposts in the branch. A dendritic branch was considered stable if its length did not change for more than 0.5 μm since our measurement error was within 0.5 μm.
Laser damaging of dendritic Golgi outposts
Early 3rd instar larvae were used for laser damaging of dendritic Golgi outposts. For efficient damaging of Golgi outposts, terminal branches that contained one or two Golgi outposts were selected. The power of 488 nm-laser was set at 15mW. After laser illumination, the larvae were transferred to a grape-juice agar plate before mounted again for imaging 5 minutes later. Images of the same dendritic region were taken before, immediately after, and 5 minutes after illumination. Laser damaging substantially reduced or eliminated GalT-EYFP, actin-GFP, or mito-GFP signal in every experiment. The length of dendritic branch immediately after illumination and 5 min after illumination was measured.
The statistical analysis of laser damaging data was carried out according to standard methods for analyzing nominal variables (Glantz, 2005). Briefly, standard error σ was calculated as [r(1− r)/n] 1/2, where r is the percentage of extended or retracted branches, and n is the total number of branches. To test for the difference between two nominal data sets, a value of z was calculated by the equation z = (r1−r2)/[r1(1−r1)/n1+r2(1−r2)/n2]1/2, where r1 and r2 are the percentage of extended or retracted branches of the two experimental groups to be compared, and n1, n2 are the total number of branches of the two groups. The probability (p) corresponding to the z value was then obtained from the standard normal distribution table.
Supplementary Material
Acknowledgments
We would like to thank Jennifer Lippincott-Schwartz for mammalian Sar1 cDNA construct, Vivek Malhotra for ManII-EGFP cDNA, Ophelia Papoulas and John Sisson for anti-Lva antibody, Jillian Brechbiel and Elizabeth Gavis for UAS-mCD8-dsRed transgenic lines, Barry Dickson for the VDRC UAS-Lva-RNAi line, and Robin Shaw for advice on FRAP experiment. We also thank Tong Cheng, Denan Wang, Kelly Borden, and Gebeyehu Ayalew for technical supports. This work is supported by a NRSA fellowship and a Pathway to Independence Award (K99MH080599) from NIH to B.Y., a graduate fellowship from Genetech, Inc and the Sandler Family Supporting Foundation to Y.Z., and NIH grants (R01NS40929 and R01NS47200) to Y.N.J.. Y.N.J. and L.Y.J. are investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- Allan VJ, Thompson HM, McNiven MA. Motoring around the Golgi. Nat Cell Biol. 2002;4:E236–242. doi: 10.1038/ncb1002-e236. [DOI] [PubMed] [Google Scholar]
- Aridor M, Guzik AK, Bielli A, Fish KN. Endoplasmic Reticulum Export Site Formation and Function in Dendrites. J Neurosci. 2004;24:3770–3776. doi: 10.1523/JNEUROSCI.4775-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas PW, Deitch JS, Black MM, Banker GA. Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc Natl Acad Sci U S A. 1988;85:8335–8339. doi: 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartheld CSv. Axonal transport and neuronal transcytosis of trophic factors, tracers, and pathogens. Journal of Neurobiology. 2004;58:295–314. doi: 10.1002/neu.10315. [DOI] [PubMed] [Google Scholar]
- Bradke F, Dotti CG. Neuronal polarity: vectorial cytoplasmic flow precedes axon formation. Neuron. 1997;19:1175–1186. doi: 10.1016/s0896-6273(00)80410-9. [DOI] [PubMed] [Google Scholar]
- Bradke F, Dotti CG. Changes in membrane trafficking and actin dynamics during axon formation in cultured hippocampal neurons. Microsc Res Tech. 2000;48:3–11. doi: 10.1002/(SICI)1097-0029(20000101)48:1<3::AID-JEMT2>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Clark IE, Jan LY, Jan YN. Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development. 1997;124:461–470. doi: 10.1242/dev.124.2.461. [DOI] [PubMed] [Google Scholar]
- Cox RT, Spradling AC. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development. 2003;130:1579–1590. doi: 10.1242/dev.00365. [DOI] [PubMed] [Google Scholar]
- Craig AM, Wyborski RJ, Banker G. Preferential addition of newly synthesized membrane protein at axonal growth cones. Nature. 1995;375:592–594. doi: 10.1038/375592a0. [DOI] [PubMed] [Google Scholar]
- Dotti CG, Simons K. Polarized sorting of viral glycoproteins to the axon and dendrites of hippocampal neurons in culture. Cell. 1990;62:63–72. doi: 10.1016/0092-8674(90)90240-f. [DOI] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwine J, Miyashiro K, Kacharmina JE, Job C. Local translation of classes of mRNAs that are targeted to neuronal dendrites. Proc Natl Acad Sci U S A. 2001;98:7080–7085. doi: 10.1073/pnas.121146698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger FP, Hughes TE, Novick P. Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell. 1998;92:559–571. doi: 10.1016/s0092-8674(00)80948-4. [DOI] [PubMed] [Google Scholar]
- Gardiol A, Racca C, Triller A. Dendritic and postsynaptic protein synthetic machinery. J Neurosci. 1999;19:168–179. doi: 10.1523/JNEUROSCI.19-01-00168.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudilliere B, Konishi Y, de la Iglesia N, Yao G, Bonni A. A CaMKII-NeuroD Signaling Pathway Specifies Dendritic Morphogenesis. Neuron. 2004;41:229–241. doi: 10.1016/s0896-6273(03)00841-9. [DOI] [PubMed] [Google Scholar]
- Glantz SA. Primer of biostatistics (McGRAW-HILL) 2005. How to analyze rates and proportions; pp. 126–178. [Google Scholar]
- Grueber WB, Ye B, Moore AW, Jan LY, Jan YN. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol. 2003;13:618–626. doi: 10.1016/s0960-9822(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Yang CH, Younger S, Borden K, Jan LY, Jan YN. Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development. 2007;134:55–64. doi: 10.1242/dev.02666. [DOI] [PubMed] [Google Scholar]
- Harada A, Teng J, Takei Y, Oguchi K, Hirokawa N. MAP2 is required for dendrite elongation, PKA anchoring in dendrites, and proper PKA signal transduction. J Cell Biol. 2002;158:541–549. doi: 10.1083/jcb.200110134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasdemir B, Fitzgerald DJ, Prior IA, Tepikin AV, Burgoyne RD. Traffic of Kv4 K+ channels mediated by KChIP1 is via a novel post-ER vesicular pathway. J Cell Biol. 2005;171:459–469. doi: 10.1083/jcb.200506005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton AC, Ehlers MD. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. The Journal of neuroscience. 2003;23:6188–6199. doi: 10.1523/JNEUROSCI.23-15-06188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton AC, Racz B, Monson EE, Lin AL, Weinberg RJ, Ehlers MD. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 2005;48:757–771. doi: 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Kupfer A, Louvard D, Singer SJ. Polarization of the Golgi apparatus and the microtubule-organizing center in cultured fibroblasts at the edge of an experimental wound. Proc Natl Acad Sci U S A. 1982;79:2603–2607. doi: 10.1073/pnas.79.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T, Pilot F. Developmental control of cell morphogenesis: a focus on membrane growth. Nat Cell Biol. 2003;5:103–108. doi: 10.1038/ncb0203-103. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Wieschaus E. Polarized insertion of new membrane from a cytoplasmic reservoir during cleavage of the Drosophila embryo. J Cell Biol. 2000;150:849–860. doi: 10.1083/jcb.150.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee T, Winter C, Marticke SS, Lee A, Luo L. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron. 2000;25:307–316. doi: 10.1016/s0896-6273(00)80896-x. [DOI] [PubMed] [Google Scholar]
- Lein P, Johnson M, Guo X, Rueger D, Higgins D. Osteogenic protein-1 induces dendritic growth in rat sympathetic neurons. Neuron. 1995;15:597–605. doi: 10.1016/0896-6273(95)90148-5. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- Morin-Ganet MN, Rambourg A, Deitz SB, Franzusoff A, Kepes F. Morphogenesis and dynamics of the yeast Golgi apparatus. Traffic. 2000;1:56–68. doi: 10.1034/j.1600-0854.2000.010109.x. [DOI] [PubMed] [Google Scholar]
- Papoulas O, Hays TS, Sisson JC. The golgin Lava lamp mediates dynein-based Golgi movements during Drosophila cellularization. Nat Cell Biol. 2005;7:612–618. doi: 10.1038/ncb1264. [DOI] [PubMed] [Google Scholar]
- Parton RG, Simons K, Dotti CG. Axonal and dendritic endocytic pathways in cultured neurons. J Cell Biol. 1992;119:123–137. doi: 10.1083/jcb.119.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce JP, Mayer T, McCarthy JB. Evidence for a satellite secretory pathway in neuronal dendritic spines. Curr Biol. 2001;11:351–355. doi: 10.1016/s0960-9822(01)00077-x. [DOI] [PubMed] [Google Scholar]
- Raab-Graham KF, Haddick PC, Jan YN, Jan LY. Activity- and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science. 2006;314:144–148. doi: 10.1126/science.1131693. [DOI] [PubMed] [Google Scholar]
- Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shi SH, Cheng T, Jan LY, Jan YN. The immunoglobulin family member dendrite arborization and synapse maturation 1 (Dasm1) controls excitatory synapse maturation. Proc Natl Acad Sci U S A. 2004;101:13346–13351. doi: 10.1073/pnas.0405371101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Simons K, Fuller SD. Cell surface polarity in epithelia. Annu Rev Cell Biol. 1985;1:243–288. doi: 10.1146/annurev.cb.01.110185.001331. [DOI] [PubMed] [Google Scholar]
- Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Reeves TM. Protein-synthetic machinery beneath postsynaptic sites on CNS neurons: association between polyribosomes and other organelles at the synaptic site. J Neurosci. 1988;8:176–184. doi: 10.1523/JNEUROSCI.08-01-00176.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Compartmentalized synthesis and degradation of proteins in neurons. Neuron. 2003;40:347–359. doi: 10.1016/s0896-6273(03)00635-4. [DOI] [PubMed] [Google Scholar]
- van Vliet C, Thomas EC, Merino-Trigo A, Teasdale RD, Gleeson PA. Intracellular sorting and transport of proteins. Prog Biophys Mol Biol. 2003;83:1–45. doi: 10.1016/s0079-6107(03)00019-1. [DOI] [PubMed] [Google Scholar]
- Velasco A, Hendricks L, Moremen KW, Tulsiani DR, Touster O, Farquhar MG. Cell type-dependent variations in the subcellular distribution of alpha-mannosidase I and II. J Cell Biol. 1993;122:39–51. doi: 10.1083/jcb.122.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TH, Polishchuk RS, Caplan S, Hirschberg K, Lippincott-Schwartz J. Maintenance of Golgi structure and function depends on the integrity of ER export. J Cell Biol. 2001;155:557–570. doi: 10.1083/jcb.200107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BS, Nuoffer C, Meinkoth JL, McCaffery M, Feramisco JR, Balch WE, Farquhar MG. A Rab1 mutant affecting guanine nucleotide exchange promotes disassembly of the Golgi apparatus. J Cell Biol. 1994;125:557–571. doi: 10.1083/jcb.125.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- Yeaman C, Ayala MI, Wright JR, Bard F, Bossard C, Ang A, Maeda Y, Seufferlein T, Mellman I, Nelson WJ, Malhotra V. Protein kinase D regulates basolateral membrane protein exit from trans-Golgi network. Nat Cell Biol. 2004;6:106–112. doi: 10.1038/ncb1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaal KJ, Smith CL, Polishchuk RS, Altan N, Cole NB, Ellenberg J, Hirschberg K, Presley JF, Roberts TH, Siggia E, et al. Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell. 1999;99:589–601. doi: 10.1016/s0092-8674(00)81548-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


