Abstract
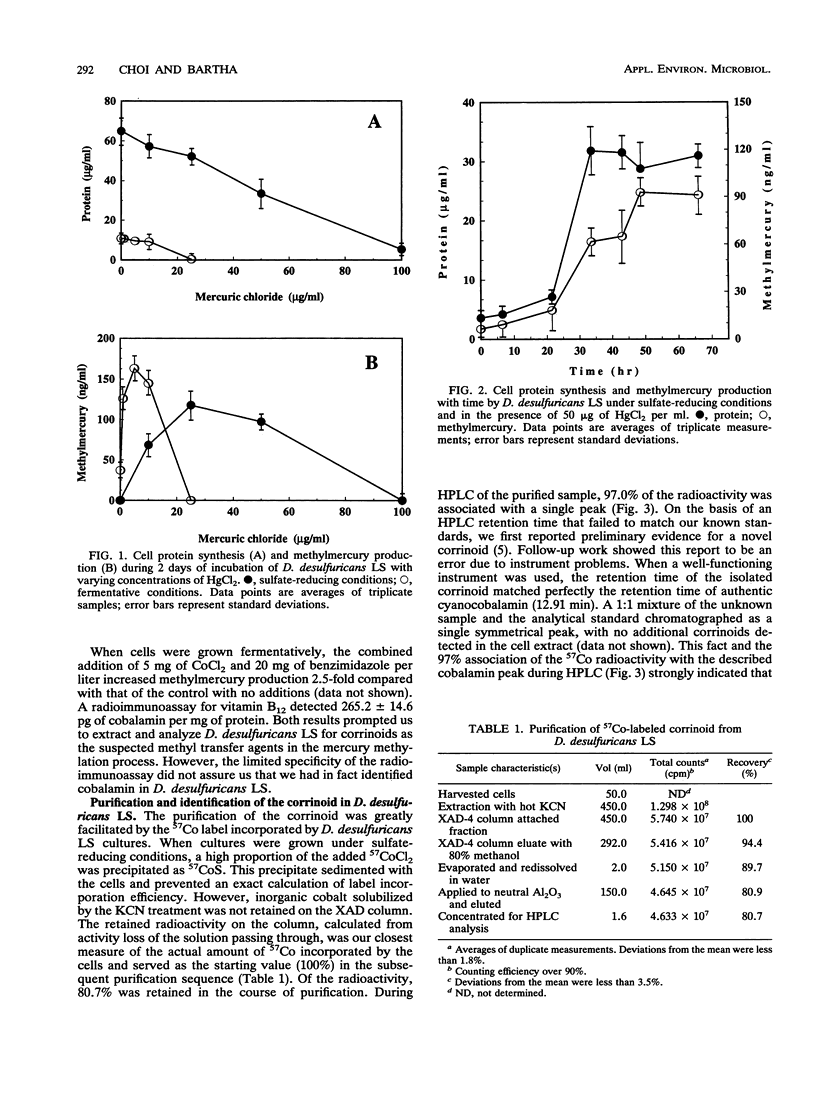
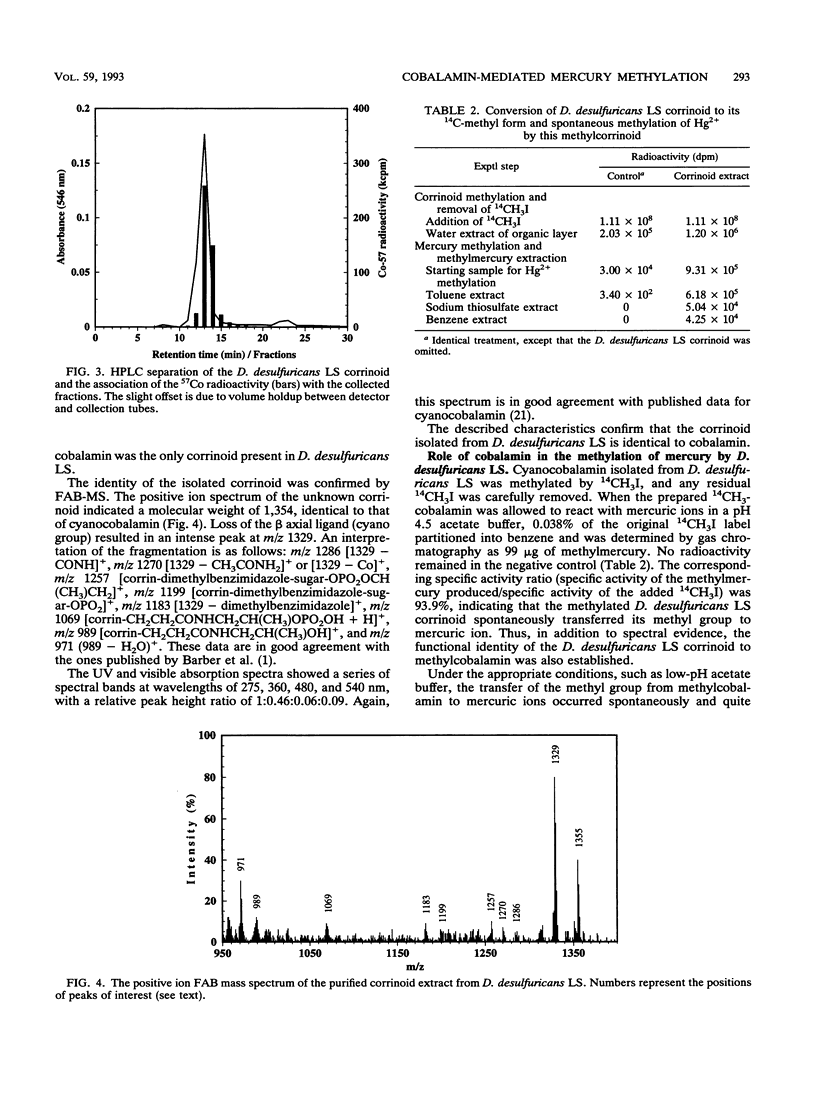
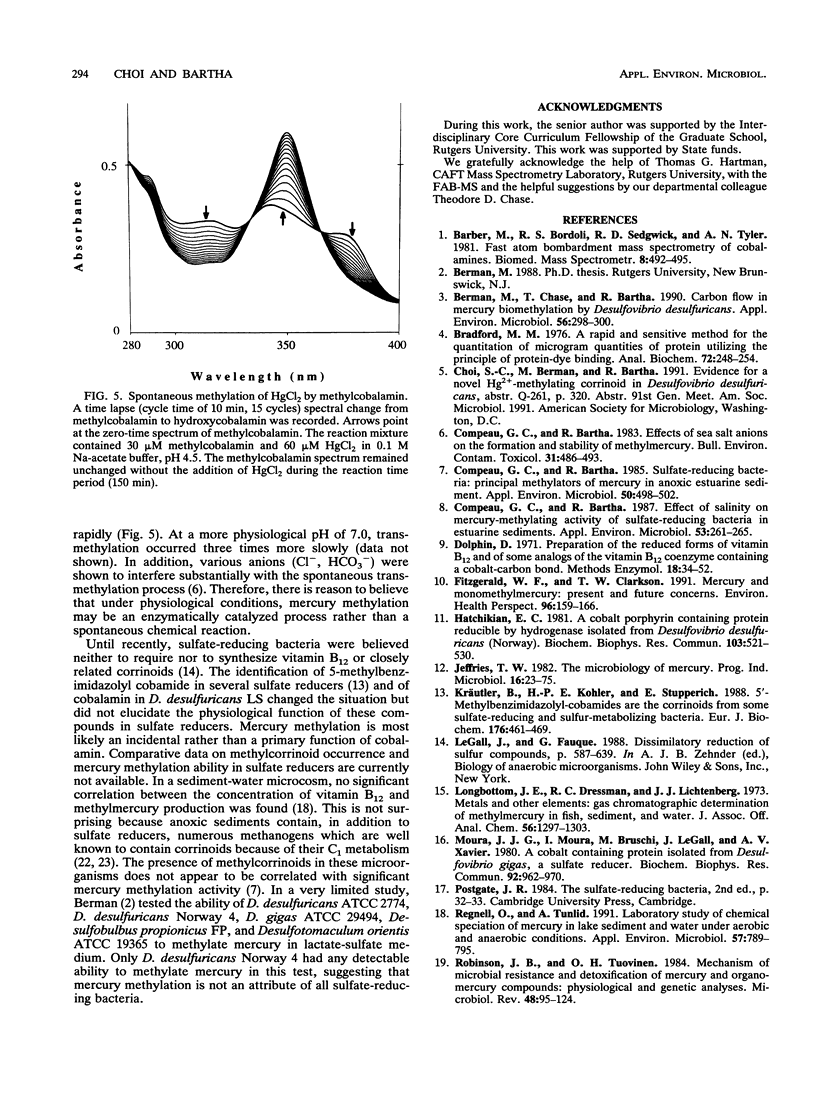
The prominence of sulfate reducers in mercury biomethylation prompted the examination of the methyl carrier and mercury methylation activity of Desulfovibrio desulfuricans LS. There was a low degree of mercury tolerance and a high degree of methylation during fermentative growth; the opposite was true during sulfate reduction. During 2 days of fermentative growth, up to 37% of HgCl2 was methylated at 0.1 micrograms/ml, but only 1.5% was methylated at 10.0 micrograms/ml. Less than 1% of the added HgCl2 was methylated under sulfate-reducing conditions. D. desulfuricans LS radioimmunoassay results were positive for cobalamin. The addition of CoCl2 and benzimidazole to fermentative cultures increased methylation activity. From D. desulfuricans LS grown in the presence of (57)CoCl2, a corrinoid was extracted and purified. High-performance liquid chromatography analysis of the purified extract yielded a single peak with the retention time of cobalamin, and 97% of the (57)Co radioactivity was associated with this peak. Fast atom bombardment and UV and visible spectra of the isolated corrinoid matched those of cobalamin. When methylated with (14)CH3I, the isolated corrinoid methylated Hg(2+) with a 93.9% preservation of (14)C specific activity. We conclude that D. desulfuricans LS methylates mercury via cobalamin (vitamin B12). Under physiological conditions, the enzymatic catalysis of this reaction is likely.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berman M., Chase T., Bartha R. Carbon Flow in Mercury Biomethylation by Desulfovibrio desulfuricans. Appl Environ Microbiol. 1990 Jan;56(1):298–300. doi: 10.1128/aem.56.1.298-300.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Compeau G. C., Bartha R. Effect of salinity on mercury-methylating activity of sulfate-reducing bacteria in estuarine sediments. Appl Environ Microbiol. 1987 Feb;53(2):261–265. doi: 10.1128/aem.53.2.261-265.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compeau G. C., Bartha R. Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediment. Appl Environ Microbiol. 1985 Aug;50(2):498–502. doi: 10.1128/aem.50.2.498-502.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compeau G., Bartha R. Effects of sea salt anions on the formation and stability of methylmercury. Bull Environ Contam Toxicol. 1983 Oct;31(4):486–493. doi: 10.1007/BF01622282. [DOI] [PubMed] [Google Scholar]
- Fitzgerald W. F., Clarkson T. W. Mercury and monomethylmercury: present and future concerns. Environ Health Perspect. 1991 Dec;96:159–166. doi: 10.1289/ehp.9196159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchikian E. C. A cobalt porphyrin containing protein reducible by hydrogenase isolated from Desulfovibrio desulfuricans (Norway). Biochem Biophys Res Commun. 1981 Nov 30;103(2):521–530. doi: 10.1016/0006-291x(81)90483-6. [DOI] [PubMed] [Google Scholar]
- Kräutler B., Kohler H. P., Stupperich E. 5'-Methylbenzimidazolyl-cobamides are the corrinoids from some sulfate-reducing and sulfur-metabolizing bacteria. Eur J Biochem. 1988 Sep 15;176(2):461–469. doi: 10.1111/j.1432-1033.1988.tb14303.x. [DOI] [PubMed] [Google Scholar]
- Moura J. J., Moura I., Bruschi M., Le Gall J., Xavier A. V. A cobalt containing protein isolated from Desulfovibrio gigas, a sulfate reducer. Biochem Biophys Res Commun. 1980 Feb 12;92(3):962–970. doi: 10.1016/0006-291x(80)90796-2. [DOI] [PubMed] [Google Scholar]
- Regnell O., Tunlid A. Laboratory Study of Chemical Speciation of Mercury in Lake Sediment and Water under Aerobic and Anaerobic Conditions. Appl Environ Microbiol. 1991 Mar;57(3):789–795. doi: 10.1128/aem.57.3.789-795.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. B., Tuovinen O. H. Mechanisms of microbial resistance and detoxification of mercury and organomercury compounds: physiological, biochemical, and genetic analyses. Microbiol Rev. 1984 Jun;48(2):95–124. doi: 10.1128/mr.48.2.95-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto M., Nakano A., Kinjo Y., Higashi H., Futatsuka M. Present mercury levels in red blood cells of nearby inhabitants about 30 years after the outbreak of Minamata disease. Ecotoxicol Environ Saf. 1991 Aug;22(1):58–66. doi: 10.1016/0147-6513(91)90047-s. [DOI] [PubMed] [Google Scholar]
- Stupperich E., Steiner I., Rühlemann M. Isolation and analysis of bacterial cobamides by high-performance liquid chromatography. Anal Biochem. 1986 Jun;155(2):365–370. doi: 10.1016/0003-2697(86)90447-1. [DOI] [PubMed] [Google Scholar]


