Abstract
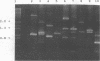
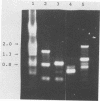
Differences in the 16S rRNA genes (16S rDNA) which can be used to discriminate Listeria monocytogenes from Listeria innocua have been detected. The 16S rDNA were amplified by polymerase chain reaction with a set of oligonucleotide primers which flank a 1.5-kb fragment. Sequence differences were observed in the V2 region of the 16S rDNA both between L. monocytogenes Scott A and L. innocua and between different L. monocytogenes serotypes. Although L. monocytogenes SLCC2371 had the same V2 region sequence as L. innocua, the two species were different within the V9 region at nucleotides 1259 and 1292, in agreement with previous studies (R.-F. Wang, W.-W. Cao, and M.G. Johnson, Appl. Environ. Microbiol. 57:3666-3670, 1991). Intraspecies discrimination of L. monocytogenes strains was achieved by using the patterns generated by random amplified polymorphic DNA primers. Although some distinction can be made within the L. monocytogenes species by their 16S rDNA sequence, a far greater discrimination within species could be made by generating random amplified polymorphic DNA patterns from chromosomal DNA. By using a number of 10-bp primers, unique patterns for each isolate which in all cases examined differentiate between various L. monocytogenes serotypes, even though they may have the same 16S rRNA sequences, could be generated.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baloga A. O., Harlander S. K. Comparison of methods for discrimination between strains of Listeria monocytogenes from epidemiological surveys. Appl Environ Microbiol. 1991 Aug;57(8):2324–2331. doi: 10.1128/aem.57.8.2324-2331.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerlin P., Piffaretti J. C. Typing of human, animal, food, and environmental isolates of Listeria monocytogenes by multilocus enzyme electrophoresis. Appl Environ Microbiol. 1991 Jun;57(6):1624–1629. doi: 10.1128/aem.57.6.1624-1629.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerlin P., Rocourt J., Piffaretti J. C. Taxonomy of the genus Listeria by using multilocus enzyme electrophoresis. Int J Syst Bacteriol. 1991 Jan;41(1):59–64. doi: 10.1099/00207713-41-1-59. [DOI] [PubMed] [Google Scholar]
- Collins M. D., Wallbanks S., Lane D. J., Shah J., Nietupski R., Smida J., Dorsch M., Stackebrandt E. Phylogenetic analysis of the genus Listeria based on reverse transcriptase sequencing of 16S rRNA. Int J Syst Bacteriol. 1991 Apr;41(2):240–246. doi: 10.1099/00207713-41-2-240. [DOI] [PubMed] [Google Scholar]
- Farber J. M., Peterkin P. I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991 Sep;55(3):476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. W., Sankoff D., Cedergren R. J. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 1984 Jul 25;12(14):5837–5852. doi: 10.1093/nar/12.14.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard P. J., Harsono K. D., Luchansky J. B. Differentiation of Listeria monocytogenes, Listeria innocua, Listeria ivanovii, and Listeria seeligeri by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1992 Feb;58(2):709–712. doi: 10.1128/aem.58.2.709-712.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger J. D., Johnson A., Croan D., Flynn P., Whippie K., Kimball M., Lawrie J., Curiale M. Comparative studies of nucleic acid hybridization assay for Listeria in foods. J Assoc Off Anal Chem. 1988 May-Jun;71(3):669–673. [PubMed] [Google Scholar]
- Loessner M. J., Busse M. Bacteriophage typing of Listeria species. Appl Environ Microbiol. 1990 Jun;56(6):1912–1918. doi: 10.1128/aem.56.6.1912-1918.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. B., Williams J. G., Tanksley S. D. Rapid identification of markers linked to a Pseudomonas resistance gene in tomato by using random primers and near-isogenic lines. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2336–2340. doi: 10.1073/pnas.88.6.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurier S. I., Audurier A., Marquet-Van der Mee N., Notermans S., Wernars K. A comparative study of randomly amplified polymorphic DNA analysis and conventional phage typing for epidemiological studies of Listeria monocytogenes isolates. Res Microbiol. 1992 Jun;143(5):507–512. doi: 10.1016/0923-2508(92)90097-8. [DOI] [PubMed] [Google Scholar]
- Mazurier S. I., Wernars K. Typing of Listeria strains by random amplification of polymorphic DNA. Res Microbiol. 1992 Jun;143(5):499–505. doi: 10.1016/0923-2508(92)90096-7. [DOI] [PubMed] [Google Scholar]
- Piffaretti J. C., Kressebuch H., Aeschbacher M., Bille J., Bannerman E., Musser J. M., Selander R. K., Rocourt J. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc Natl Acad Sci U S A. 1989 May;86(10):3818–3822. doi: 10.1073/pnas.86.10.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wang R. F., Cao W. W., Johnson M. G. Development of a 16S rRNA-based oligomer probe specific for Listeria monocytogenes. Appl Environ Microbiol. 1991 Dec;57(12):3666–3670. doi: 10.1128/aem.57.12.3666-3670.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991 Jan;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley I. V., Ashton F. Restriction enzyme analysis of Listeria monocytogenes strains associated with food-borne epidemics. Appl Environ Microbiol. 1991 Apr;57(4):969–975. doi: 10.1128/aem.57.4.969-975.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann M., Czajka J., Barany F., Batt C. A. Discrimination of Listeria monocytogenes from other Listeria species by ligase chain reaction. Appl Environ Microbiol. 1992 Nov;58(11):3443–3447. doi: 10.1128/aem.58.11.3443-3447.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]