Abstract
Nitric oxide (NO) induction through the inducible NO synthase has been demonstrated to cause cell death in macrophages. We demonstrate that, in macrophages that have been rendered resistant to apoptosis induced by inducible NO synthase (RES cells), exposure to exogenous NO donors results in a hypersensitive apoptosis reaction when compared with the parental RAW 264.7 cells. The apoptosis induced via exogenous NO donors was found to be caspase 3-independent. Although caspase 3 activity was stimulated in the apoptotic macrophages, inhibition of caspase 3 by the inhibitor DEVD-CHO (N-acetyl-Asp-Glu-Val-Asp-aldehyde) did not reverse the apoptosis induced by the NO donor S-nitrosoglutathione (GSNO). This suggests that although caspase 3 activity is stimulated during apoptosis in macrophages, this signal is not sufficient to induce apoptosis. Cleavage of the enzyme poly(ADP ribose) polymerase mirrors our results of the caspase activity. Interestingly, we show that exogenous NO donation results in an accumulation of cells at the G2/M-phase border. Here, we demonstrate that the mitogen activated protein kinase kinase (MEK) inhibitor PD 098059 can be used to reverse the G2/M-phase block and show that this treatment also inhibits the observed apoptosis in RES macrophages. Treatment with the MEK inhibitor also reversed both the caspase 3 activity and poly(ADP ribose) polymerase cleavage in cells treated with GSNO. This result indicates that the mitogen-activated protein kinase pathway may be involved in regulation of the caspase cascade. Alternatively, it may suggest an activity for the MEK inhibitor heretofore not observed, that of a cyclin kinase inhibitor. Our results suggest that selection of macrophages by resistance to endogenously generated NO may cause hypersensitivity to exogenous NO donors. These findings have relevant implications for the treatment of apoptotic-resistant cell populations that may occur in both cancer and atheroma.
Programmed cell death (apoptosis) occurs in many animal tissues during development (1) and is necessary to eliminate unwanted host cells and achieve homeostasis (2). The term apoptosis describes a tightly regulated process of cell death characterized by plasma membrane blebbing, chromatin condensation, loss of cell volume, and DNA fragmentation (3, 4). Studies with the nematode Caenorhabditis elegans have shown that specific genes are necessary for apoptosis to occur, like ced-3 and ced-4, whereas other genes such as ced-9 protect from cell death (5, 6). In mammals, ced-3 homologs have been described as a family of at least 10 cysteine proteases with the specificity for cleavage after an aspartate residue. All members of the ced-3 family are found as inactive zymogens that become activated by proteolytic cleavage to the active dimeric or tetrameric species (7). These cysteine proteases, formerly known as the ICE (interleukin 1β converting enzyme) family, are now called caspases and can be divided into three subclasses, based on sequence homology (8): the ICE (caspase 1)-like family, the CPP32 (caspase 3)-like family, and the ICH-1 (caspase 2)-like family (9, 10). Recently, several target proteins have been identified that are cleaved by caspases during the apoptosis process including lamin A, lamin B1, poly(ADP ribosyl) polymerase (PARP), topoisomerase I, sterol regulatory element-binding protein, actin, the retinoblastoma protein, D4-GDP dissociation inhibitor, and a 70-kDa component of the U1 splicing particle (11). To date, however, very little is known about the activation process for caspases. Many types of stimuli such as the Fas ligand, staurosporine, actinomycin D, etoposide, radiation, or nitric oxide (NO) can activate caspase cascade, leading to apoptosis (12).
NO is produced by the enzyme NO synthase (NOS) that catalyzes the oxidation of l-arginine yielding NO and l-citrullin (13). NOS exists in three isoforms, the constitutive neuronal and endothelial forms, which produce low levels of NO, and the inducible NOS (iNOS; in macrophages), which produces high amounts of NO (14). Production of NO normally leads to activation of the guanylate cyclase, which transforms GTP into cGMP, a second messenger that activates specific protein kinases (15). NO, because of a high redox potential, is also able to modify enzymes directly (16, 17). These cGMP-independent mechanisms are more closely associated with nitrosation, nitration, and oxidation (18–20).
Stimulation of the mouse macrophage cell line RAW 264.7 with lipopolysaccharide (LPS)/interferon γ (IFN-γ) induces the activation of iNOS and results in cell death. This is reversed by the NOS inhibitor N(G)-monomethyl-l-arginine (21). These results indicated that formation of endogenous NO is involved in the LPS/IFN-γ-induced cell death in macrophages. Our laboratory has developed a new cell line resistant to LPS/IFN-γ-induced cell death (RES cells). RES cells were derived from the mouse macrophage cell line RAW 264.7 by repeated exposure to LPS/IFN-γ, followed by outgrowth of viable cells. Characterization of RES cells has shown that the amount of superoxide production and heat shock proteins are increased (22). Because change in NO levels led to apoptosis in RAW cells, we sought to determine whether the adaptation of RES cells was because of changes in the NO response. Interestingly, when RES cells were treated with the NO donor S-nitrosoglutathione (GSNO), we observed remarkable apoptosis. Therefore, the present study is focused on the hypersensitivity of the RES cells to exogenous NO and how apoptosis is mediated during this response.
MATERIALS AND METHODS
Materials.
Leupeptin, phenylmethylsulfonyl fluoride, glutathione, sodium nitrite, DTT, and Hepes were from Sigma. Pepstatin A and CHAPS were from Fluka; DEVD-AFC (N-acetyl-Asp-Glu-l-Val-Asp-7-amino-4-trifluoromethyl coumarin, a caspase 3 fluorogenic substrate), YVAD-AFC (a caspase 1 fluorogenic substrate; where YVAD is Tyr-Val-Ala-Asp), DEVD-CHO (a caspase 3 inhibitor; where CHO is aldehyde), YVAD-CHO (a caspase 1 inhibitor), and anti-PARP antibody were from Biomol. The mitogen-activated protein kinase kinase (MEK) inhibitor PD 098059 was a gift from Parke Davis. ECL (enhanced chemiluminescence) reagents were from Amersham. Anti-rabbit IgG, horseradish peroxidase-conjugate was from Bio-Rad. RPMI 1640 medium and cell culture supplements were from GIBCO/BRL/Life Technology. Fetal bovine serum was from HyClone.
Cell Culture.
The mouse macrophage cell lines RAW 264.7 and RES were maintained in RPMI 1640 medium supplemented with penicillin (100 units/ml), streptomycin (100 μg/ml), 2 mM glutamine, and 10% (vol/vol) heat-inactivated fetal bovine serum (complete RPMI). All experiments were performed with complete RPMI.
Preparation of Cell Lysates.
For each assay, 4 × 106 cells were incubated in 100-mm dishes with various concentrations of GSNO for 8 h or as indicated. Cells were scraped and centrifuged at 500 × g, at 4°C. The pellets were resuspended in 200 μl of lysate buffer [100 mM Hepes, pH 7.5/10% sucrose/0.1% CHAPS/1 mM EDTA/10 mM DTT containing the protease inhibitors 1 mM phenylmethylsulfonyl fluoride, pepstatin (10 μg/ml), and leupeptin (10 μg/ml)]. The cellular material was left on ice for 30 min and then sonicated for 10 s at a 10% pulse with a Heart System ultrasonic processor XL. The lysates were centrifuged at 9,000 × g for 7 min at 4°C. The supernatants were frozen at −20°C. Protein concentration of the supernatants was quantified with the Bradford (Bio-Rad) protein assay. For inhibitor studies, 4 × 106 cells were preincubated with 10 μM DEVD-CHO (caspase 3 inhibitor) or 50 μM MEK inhibitor (PD 098059) for 1 h, before addition of GSNO for 8 h.
Mitogen-Activated Protein Kinase (MAPK) Kinase Assay.
Kinase activity assays were performed by using the nonradioactive kinase assay kit from New England Biolabs, according to the manufacturer’s instructions. Briefly, cells were incubated for 18 h in serum-free medium before treatment with the described activators for the indicated periods. After stimulation, the cells were washed with ice-cold PBS containing 1% Na3VO4. After washing, the cells were lysed in the tissue culture dishes with 1 ml of lysis buffer [20 mM Tris⋅HCl, pH 7.5/150 mM NaCl/1 mM EDTA/1 mM EGTA/1% Triton X-100/2.5 mM sodium pyrophosphate/1 mM β-glycerophosphate/1 mM sodium vanadate/leupeptin (1 μg/ml)] for 5 min on ice. The lysis mixture was sonicated (Heat Systems Ultrasonics, Farmingdale, NY) for four 5-s periods at the maximal micro-tip setting (3). After sonication, the sample was centrifuged for 10 min at 8,000 × g at 4°C, and the soluble supernatant fraction was removed and stored at −80°C. Protein concentrations were determined by using the bicinchonic acid assay (BCA, Pierce). MAPK was immunoprecipitated by incubation of 500 μg of macrophage lysate with specific antibodies toward phospho-MAPK followed by immunoprecipitation with protein A-Sepharose and then pelleted by centrifugation in a microcentrifuge for 30 s. The supernatant was removed and the pellet was washed once with lysis buffer and three times with kinase buffer. After washing, the pellet was suspended in 50 μl 1× kinase buffer with 100 μM ATP and the MAP kinase target protein Elk-1. After 30 min at 27°C, the reaction was terminated and the mixture was separated on a 12.5% SDS/PAGE gel. Western blotting was performed by using anti-phosphospecific Elk-1 antibodies.
Cleavage of Fluorogenic Substrate.
Lysates (50 μg of protein, prepared as described above) were incubated for 60 min at 32°C in 100 mM Hepes, pH 7.5/10% sucrose/0.1% CHAPS/1 mM EDTA/10 mM DTT containing the protease inhibitors 1 mM phenylmethylsulfonyl fluoride, pepstatin (10 μg/ml), and leupeptin (10 μg/ml) with the fluorogenic substrate DEVD-AFC (25 μM) in a total volume of 500 μl. Cleavage of the substrate emitted a fluorescence signal that was quantified in a Perkin–Elmer LS3 fluorescence spectrophotometer (excitation, 400 nm; emission, 505 nm).
Synthesis of GSNO.
Glutathione was dissolved in 2 M HCl at 4°C followed by addition of equimolar amount of sodium nitrite (NaNO2). The mixture was stirred for 40 min at 4°C. After addition of 10 ml of acetone, precipitates were filtered, washed three times with acetone and ether, and dried under vacuum. The resulting GSNO displays absorption maxima at 335 and 545 nm with extinction coefficients of 922 and 15.9 dl2 per mol per cm, respectively. GSNO solutions were prepared just before the experiment.
PARP Western Blot Analysis.
Whole-cell lysates (50 μg) were separated on an 8% reducing polyacrylamide gel and then transferred to nitrocellulose membranes. The membranes were washed three times with PBS/0.1% Tween 20 and blocked in 5% milk powder/PBS/0.1% Tween 20 for 2 h at room temperature, followed by the incubation with a polyclonal antibody against PARP at a 1:4,000 dilution overnight at 4°C. After washing the blots five times with PBS/0.1% Tween 20, the membranes were incubated with horseradish peroxidase-conjugated anti-rabbit IgG antibody in blocking buffer for 1 h. After washing, the proteins were visualized by using the ECL substrate system.
Measurement of Apoptosis.
After incubation with GSNO, 4 × 106 cells were scraped and centrifuged at 500 × g at 4°C. The pellets were washed once with 5 ml of ice-cold PBS, then permeabilized in 1 ml of 80% ice-cold ethanol, and left on ice for 30 min. Cells were centrifuged at 500 × g at 4°C for 5 min, washed with 1 ml of ice-cold PBS, and stained with propidium iodide (50 μg/ml) in PBS/0.1% Triton X-100/0.1 mM EDTA/50 μg of RNase overnight at 4°C. Apoptosis measurement was carried out on an ELITE profile fluorescence-activated cell sorter (Coulter) using a cell cycle analysis doublet discrimination protocol.
RESULTS
Caspase 3 Activity in RAW and RES Cells Induced by GSNO.
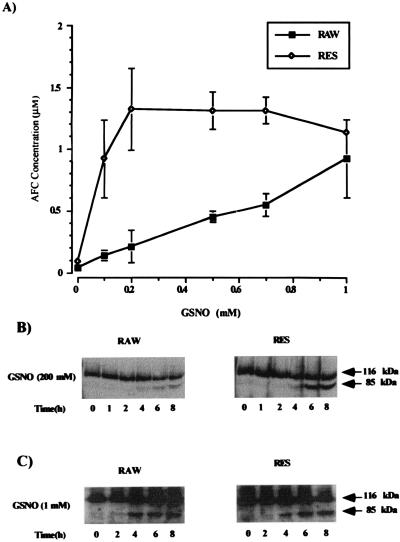
In this study, we examine the effect of GSNO on caspase 3 activity in RAW and RES cells. Because caspases are involved in the execution of apoptosis, protease activity was measured in RAW and RES cells by following the cleavage of the caspase 3 substrate DEVD-AFC in the cytosolic fraction of GSNO-treated cells (Fig. 1A). Active caspase 3 cleaves the substrate after the second Asp residue and releases the fluorescent fragment (7-amino-4-trifluoromethyl coumarin or AFC) from the DEVD-AFC substrate. The accumulation of the fluorescent dye was quantitated and correlated with enzyme activity. GSNO activated caspase 3 in RAW cells in a concentration-dependent manner. Protease activity was significantly elevated at concentrations of GSNO greater than 500 μM. In RES cells, however, addition of 200 μM GSNO was enough to reach maximal caspase 3 activity in these cells. Higher concentrations of GSNO did not increase caspase 3 activity further. At 1 mM GSNO both cell lines showed nearly the same amount of caspase 3 activity. The effect of DTT on the caspase measurements was assessed; when DTT was not added to the caspase activity assay, the levels of response were decreased but the absolute ratios of activation remained the same (data not shown). The protein levels of caspase 3 in RAW and RES cells were equivalent.
Figure 1.
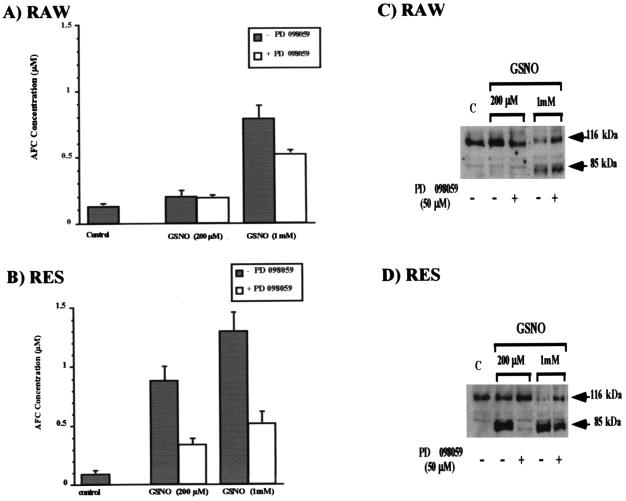
Concentration-dependent activation of caspase 3 activity by GSNO and subsequent time-dependent PARP cleavage in RAW and RES cells. (A) RAW and RES were treated for 8 h with the indicated concentrations of GSNO. Cell extracts (50 μg) were then incubated in the presence of the fluorescent caspase 3 substrate DEVD-AFC (25 μM) for 1 h at 32°C. Caspase 3 activity was measured fluorometrically after substrate cleavage with excitation at 400 nm and emission at 505 nm. Values are the mean ± SD of four experiments. (B and C) PARP cleavage was detected by Western blot analysis using a polyclonal antibody specific for PARP. The arrows indicate the 116-kDa PARP protein and its 85-kDa cleavage product. Time-dependent PARP cleavage after incubation of RAW and RES cells with either 200 μM GSNO (B) or 1 mM GSNO (C) is shown. Results are representative of three experiments.
Incubation of the cytosolic fractions from GSNO-treated RAW and RES cells with the caspase 1-specific substrate YVAD-AFC (N-acetyl-Tyr-Val-Ala-Asp-7-amino-4-trifluoromethyl coumarin) did not produce any fluorescent signal, indicating that caspase 1 is not activated in these cells during NO-induced apoptosis (data not shown).
PARP Cleavage in GSNO-Treated RAW and RES Cells.
PARP is a target protein for caspase 3 that is cleaved during the apoptotic process. Treatment of RAW and RES cells with a low concentration of GSNO (200 μM) led, in RES cells, to the cleavage of the 116-kDa PARP molecule to its 85-kDa fragment, as determined by Western blot analysis using a polyclonal antibody against PARP (Fig. 1B). PARP cleavage in RES cells occurred between 4 and 6 h, whereas little PARP cleavage could be seen in the RAW cells even after 6–8 h. At 1 mM GSNO, PARP cleavage after 4 h is similar in RAW and RES cells (Fig. 1C). These results were consistent with the measurement of caspase 3 activity under the same conditions. Caspase 3 became activated in RES cells after 4 h of treatment with 200 μM GSNO, whereas RAW cells showed very little caspase 3 activity. High concentration of GSNO (1 mM) caused caspase 3 activation in RAW and RES cells after 3–4 h of treatment (data not shown).
Induction of Apoptosis in RAW and RES Cells After GSNO Treatment.
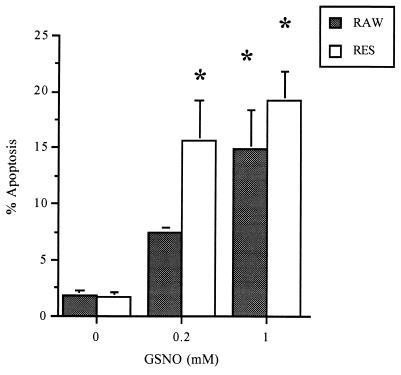
The measurement of apoptosis with flow cytometry analysis demonstrated that RES cells were more sensitive to GSNO treatment (Fig. 2). In RES cells, 200 μM or 1 mM GSNO caused 15% or 20–28% apoptosis, respectively. RAW cells showed a significant amount of apoptosis (15%, P ≤ 0.001) only at 1 mM GSNO (Fig. 2). Preincubation of both cell lines with the caspase 3 inhibitor DEVD-CHO (10 μM) followed by the addition of 200 μM or 1 mM GSNO, respectively, completely blocked the GSNO-induced caspase 3 activity (Table 1). Apoptosis was reduced 20–30% in RAW and RES cells but was not fully inhibited. Concentrations of the caspase 3 inhibitor DEVD-CHO up to 100 μM did not further decrease the amount of apoptosis. The caspase 1 inhibitor YVAD-CHO had neither an effect on caspase 3 activity nor on apoptosis (data not shown).
Figure 2.
GSNO-induced apoptosis in RAW and RES cells. Cells were treated for 8 h with 200 μM or 1 mM GSNO, respectively. Apoptosis was quantified by flow cytometry using propidium iodide exclusion. Approximately 10,000 cells were sampled, and the percentage of cells undergoing apoptosis was calculated. Data are representative of results from at least six similar experiments.
Table 1.
AFC and apoptosis values for cells treated with GSNO with and without the caspase 3 inhibitor DEVD-CHO
| Cell | Treatment | AFC μM | Apoptosis % |
|---|---|---|---|
| RAW | Control | 0.043 ± 0.02 | 1.9 ± 0.3 |
| GSNO (200 μM) | 0.21 ± 0.13 | 7.5 ± 1.4 | |
| GSNO (200 μM) + DEVD-CHO (10 μM) | 0.08 ± 0.04 | 5.8 ± 1.2 | |
| GSNO (1 mM) | 0.92 ± 0.32 | 14.2 ± 2.6 | |
| GSNO (1 mM) + DEVD-CHO (10 μM) | 0.08 ± 0.06 | 9.9 ± 2.4 | |
| RES | Controlc | 0.087 ± 0.01 | 1.7 ± 0.4 |
| GSNO (200 μM) | 1.32 ± 0.33 | 15.7 ± 3.6 | |
| GSNO (200 μM) + DEVD-CHO (10 μM) | 0.06 ± 0.04 | 12.5 ± 1.3 | |
| GSNO (1 mM) | 1.13 ± 0.01 | 19.2 ± 3.4 | |
| GSNO (1 mM) + DEVD-CHO (10 μM) | 0.07 ± 0.01 | 15.6 ± 2.4 |
Apoptosis was calculated from intercalation of propidium iodide into DNA from ethanol-fixed RAW or RES cells that had been treated with the indicated amounts of GSNO and DEVD-CHO. Control values were derived from untreated cell populations.
Kinase Activation After Treatment with 200 μM GSNO.
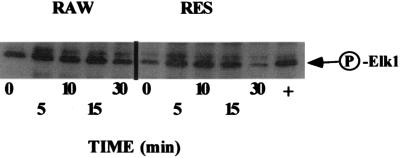
To determine whether kinase activation occurred after treatment with GSNO, we next examined MAPK activation in cells treated with GSNO. As shown in Fig. 3, MAPK is activated after GSNO treatment in both RAW and RES cells. The peak increase of MAPK activity is between 5 and 10 min, and the basal level of MAPK is higher in RAW cells than in the RES cells. The more rapid decline of MAPK in RES cells may indicate that the level of MAPK is important to inducing apoptosis. This may be especially important if the activity of the stress-activated protein kinase (SAPK) is constant. We have extended the time course for MAPK activation up to 12 h, and during this period, have noted that although the MAPK activity decreases at the 30 min time point, the activation of MAPK is sustained until 6 h after GSNO treatment (data not shown).
Figure 3.
RAW and RES cells were incubated in serum-free RPMI 1640 medium for 18 h before treatment with 200 μM GSNO for the times indicated. MAPK activity is indicated by phosphorylation of Elk-1. The level of MAPK increases in both RAW and RES cells, although the basal level in RAW cells is higher than that of the RES cells. Although the basal levels differ, the maximal response in MAPK activity appears to peak between 10 and 15 min and then decline thereafter.
Analysis of Apoptosis, Cell Cycle Progression, and the Effect of the MEK Inhibitor PD 098059 in Macrophages by Flow Cytometry.
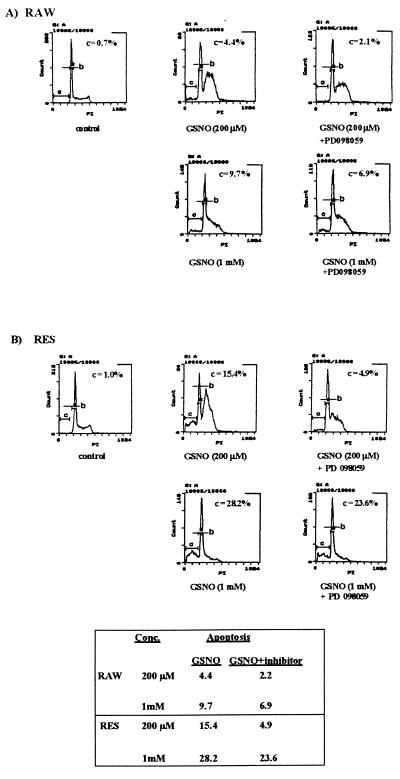
Flow cytometry analysis of apoptosis is shown in Fig. 4. Propidium iodide uptake in apoptotic cells, cells to the left of the G0/G1 peak, in Fig. 4 is reduced because of the inability of the dye to intercalate into condensed DNA during apoptosis. Cells marked with bar B are in the G0/G1 phase of the cell cycle. The peak to the right of bar B represents cells undergoing proliferation (S or G2/M phase of cell cycle) and they are, therefore, able to bind more propidium iodide because of their higher DNA (n = 2–4) content. The flow cytometry analysis of untreated RAW cells showed that most cells have been in the G0/G1 phase, some have been in a proliferating phase, and only a very small amount of apoptotic cells (0.7% apoptosis) was detected (Fig. 4A). Treatment of RAW cells with 200 μM GSNO led to G2/M-phase arrest of the cells without increasing the amount of apoptosis significantly (4.4% apoptosis).
Figure 4.
Propidium iodide staining of RAW and RES cells after treatment with GSNO in the presence or absence of the MEK inhibitor PD 098059. The cell cycle profile of RAW (A) or RES (B) cells after treatment with 200 μM or 1 mM GSNO, respectively, in the presence or absence of the MEK inhibitor PD 098059 (50 μM) is shown. After an 8-h incubation, the amount of apoptotic cells was measured by flow cytometry analysis. Bars: C, apoptotic cells; B, cells in the G0/G1 phase of the cell cycle. The peak to the right of bar B represents cells undergoing proliferation (S or G2/M phase). Data are representative of results from three experiments.
In an attempt to induce apoptosis in RAW cells treated with 200 μM GSNO, we preincubated the RAW cells with the specific MEK inhibitor PD 098059 followed by the GSNO (200 μM) treatment. By blocking the MAPK signaling pathway, we hoped to favor the stress-activated kinase response pathway. Interestingly, the incubation of the cells with GSNO (200 μM) and the MEK-specific inhibitor PD 098059 (50 μM) did not cause apoptosis but, remarkably, suppressed the G2/M-phase arrest induced by GSNO. Addition of the inhibitor also caused MEK inhibition in both RAW and RES cells (data not shown). The amount of apoptotic cells were slightly reduced (2.1% apoptosis). Incubation of the cells with 1 mM GSNO caused apoptosis (9.7%) but did not drive the cells into G2/M arrest. Treatment of RAW cells with 1 mM GSNO in the presence of the MEK inhibitor PD 098059 (50 μM) reduced apoptosis about 30% (Fig. 4A).
The cell cycle profile of untreated RES cell was similar to that of RAW cells (Fig. 4B). Only 1% apoptotic cells were detected. Incubation of RES with 200 μM GSNO increased the amount of cells arrested in G2/M phase as shown for the RAW cells. Compared with the parental RAW cells, the RES cells appear to arrest in late S phase. However, treatment of RES cells with 200 μM GSNO caused a significant amount of apoptosis (15.4%), which could be dramatically reduced by the MEK inhibitor PD 098059 (60% inhibition), as shown in Fig. 4B. PD 098059 also decreased the number of RES and RAW cells arrested in late S or G2/M phase of the cell cycle. Higher concentrations of GSNO (1 mM) increased apoptosis (28.2% apoptosis) in RES cells without inducing an S–G2/M-phase arrest. The effect of PD 098059 on RES cells treated with 1 mM GSNO was less than shown for cells treated with 200 μM GSNO. Apoptosis was only reduced approximately 10% (Fig. 4B).
NO activates the guanylate cyclase producing cGMP. To examine whether low concentrations of GSNO (200 μM) arrest RAW and RES cells in a G2/M phase via a cGMP-dependent pathway, we incubated both cell lines with the cGMP analog 8-bromo-cGMP. 8-Bromo-cGMP at 100 μM and 1 mM did not induce G2/M arrest or cause apoptosis in either RAW or RES cells. Caspase 3 activity was also not induced when the cGMP analog was used instead of GSNO (data not shown).
Effect of PD 098059 on Caspase 3 Activity Induced by GSNO.
Because the specific MEK inhibitor PD 098059 reduced GSNO-induced apoptosis in RAW and RES cells, the influence of this inhibitor on caspase 3 activity was studied further (Fig. 5). As described above, only high concentrations of GSNO (1 mM) induced caspase 3 activity in RAW cells. This protease activity could be reduced by incubating the cells with GSNO in the presence of 50 μM PD 098059 (Fig. 5A). Treatment of RES cells with 200 μM or 1 mM GSNO, respectively, in the presence of PD 098059 (50 μM) remarkably reduced caspase 3 activity (Fig. 5B).
Figure 5.
Influence of PD 098059 on the GSNO-induced caspase 3 activity and PARP cleavage in RAW and RES cells. RAW (A) and RES (B) cells were treated for 8 h with various concentrations of GSNO in the presence or absence of the MEK inhibitor PD 098059 (50 μM). Cell extracts (50 μg) were then incubated with the caspase 3 substrate DEVD-AFC (25 μM) for 1 h at 32°C. Caspase 3 activity was measured. PARP cleavage was detected by Western blot analysis using a polyclonal antibody specific for PARP. The arrows indicate the 116-kDa PARP protein and its 85-kDa cleavage product. PARP cleavage in RAW (C) or RES (D) cells is shown after incubation of the cells for 8 h with 200 mM or 1 mM GSNO, respectively, in the presence or absence of the MEK inhibitor PD 098059 (50 mM). Data are representative of results from three experiments.
To exclude a direct inhibitory effect of PD 098059 on caspase 3, we treated RES cells with 200 μM and 1 mM GSNO, respectively. Under these conditions caspase 3 was activated as described above. We incubated the cytosolic fraction of the GSNO-treated cells with DEVD-AFC in the presence or absence of 50 μM PD 098059. Addition of PD 098059 did not inhibit caspase 3 activity in vitro (data not shown).
Effect of PD 098059 on PARP Cleavage After GSNO Treatment of RAW and RES Cells.
We also examined whether preincubation with PD 098059 (50 μM) prevented PARP cleavage in RAW and RES cells induced by GSNO. Fig. 5C shows that PD 098059 was able to partially reverse PARP cleavage after an 8-h treatment with 1 mM GSNO. As described above, 200 μM GSNO did not lead to PARP cleavage in RAW cells (Fig. 5C). In RES cells, PD 098059 nearly inhibited PARP cleavage induced by 200 μM GSNO (Fig. 5D). However, PD 098059 was not able to fully prevent PARP cleavage in RES cells after an 8-h exposure to 1 mM GSNO.
DISCUSSION
Our results indicate that exogenously supplied NO, such as that derived from the NO donor GSNO, and endogenously derived NO, such as that derived via the iNOS, can have different phenotypic consequences on macrophages. Brüne and coworkers (22–24) and other laboratories (25, 26) have demonstrated that the treatment of RAW264.7 macrophages with bacterial LPS in combination with the cytokine IFN-γ resulted in apoptosis. In this study, we have demonstrated that when RAW264.7 cells are treated with exogenous NO donors, these cells are not induced to undergo apoptosis at lower levels of the NO donors and only begin to undergo apoptosis upon treatment of the cells with very high levels of the NO donor. This suggests that the signal transduction pathways responsible for initiating the apoptotic pathway are stimulated differently by iNOS as compared with NO provided from an NO donor.
Further, by using a cell line that we have previously characterized as resistant to LPS/IFN-γ-mediated cell death (RES cells), we demonstrate herein that cells that have become resistant to iNOS-induced apoptosis signaling are hypersensitive to exogenous sources of NO and that rapid apoptosis is induced in response to even low levels of the exogenous NO donor. We have characterized the apoptosis induced in the RES cells via exogenous NO treatment and have shown that caspase 3 activity is greatly increased in the RES cells as compared with the RAW cells treated with 200 μM GSNO. These results agree with reports suggesting that NO may mediate apoptosis via caspase activity (27). We observed that the half-life of the GSNO in medium was less than 1.5 h (unpublished observation), suggesting that at the time that caspase 3 cleavage occurs (after 4 h), the GSNO is no longer present to inhibit the active caspase 3 formation. Because proform caspase is not affected by GSNO, induction of apoptosis through caspase 3 formation is possible.
It has been shown that NO leads to inhibition of the cysteine proteases in vitro. Conversely, in vivo NO was able to activate and to inhibit caspase activity (12, 28–31). In our system using RAW and RES macrophages, we can examine the response of macrophages to exogenous NO that results in apoptosis and caspase 3 activation. Simultaneously, we can examine the resistance to apoptosis (RES cells) stimulated also through induction of endogenous NO, via LPS/IFN-γ activation. We also have examined the caspase activity in the RAW264.7 macrophage line in response to NO treatment. Perhaps during the selection of the RES cells from RAW cells via repeated treatment with LPS/IFN-γ, apoptotic signaling pathways were changed because of S-nitrosylation and/or nitration of proteins, thereby protecting the RES cells from apoptosis. However, if the RES cells are treated with exogenous NO, then apoptosis is enhanced, suggesting that NO may operate on different levels depending on the source of NO. The levels of iNOS produced in RAW and RES cells is equivalent, and measurement of nitrite in the two cell lines also suggests that the level of endogenous NO are the same (21). Evidence supporting this hypothesis also comes from reports where pretreatment of cells with low levels of GSNO render the cells resistant to further treatment with higher levels of GSNO (32). This also may be reflected in the changes we observe in apoptosis based on the concentration of GSNO applied. This suggests that the protection normally afforded the RAW cells to exogenously provided NO can be overcome if high enough levels of NO are used. Indeed, with a treatment of 1 mM NO, we have found that the pattern of cell cycle changes differs from that observed using lower levels of NO, potentially contributing to the difference in apoptosis observed. It is likely that at high level of GSNO, direct DNA damage may occur stimulating p53 production and cell death as has been described (32).
PARP cleavage, which has been correlated with caspase 3 activity, after treatment of RAW and RES cells with 200 μM GSNO agrees with the caspase activity pattern. Namely, PARP is cleaved faster and more completely in RES cells than in RAW cells. When the concentration of GSNO is increased to 1 mM, the cleavage of PARP is more equivalent between the two cell lines. The caspase-directed cleavage of PARP may not account for the apoptosis observed in these cells. When the caspase 3-specific inhibitor DEVD-CHO is added to RAW and RES cells (see Table 1), the caspase activity is inhibited in these cells with 200 μM and 1 mM GSNO. However, the apoptosis observed after caspase inhibitor treatment is not significantly lowered by inhibiting the caspase activity. This suggests that more that one mechanism must be responsible for inducing the apoptosis observed with exogenous NO donors, and although caspase(s) are involved, they are not the sole mechanism responsible for cell death. Dual effects for caspases have been previously proposed as an explanation for their contribution to apoptosis (33).
With low levels of GSNO, we demonstrate that both RAW and RES cells begin to accumulate in late S or early G2/M phase of the cell cycle. Other reports have also demonstrated that exogenous NO donors can lead to this type of cell cycle arrest (34–37). However, in the previous reports, the cells were not induced to undergo apoptosis and, therefore, resembled what we observed in the RAW cell population but not necessarily in the RES cell population. In contrast to what has usually been observed for cell cycle arrest, it appears that in the RES cells, the inhibition at the G2/M stage of the cycle is correlated with apoptosis via exogenous NO treatment.
Both RAW and RES cells activated the MAPK pathway in response to 200 μM GSNO. The decline of the MAPK response in RES cells appears to occur more rapidly than in the RAW cells, suggesting that MAPK may be important in protecting cells from apoptosis. However, at increased times, the MAPK response remains elevated above basal levels. It has not been determined why selection via LPS/IFN-γ makes the RES cells hypersensitive to NO donors. One potential explanation is that the activation that occurs increases SAPK endogenous signaling. Increased levels of the SAPK may “prime” these cells toward apoptosis. When a stress signal is activated, such as via the NO mediator GSNO, a lower level of MAPK may result, and therefore, the up-regulated SAPK response may be enough of a signal to begin the apoptotic process. Although the decrease in MAPK may be slight, in an environment where SAPK is increased, the balance may be shifted quite rapidly, thereby favoring apoptosis.
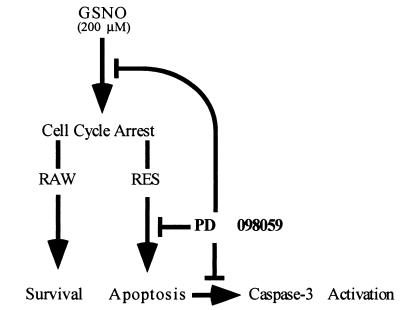
Interestingly, if the RES cells are incubated with the specific MEK inhibitor PD 098059 before exposure to the exogenous NO donor GSNO, both the G2/M cell cycle arrest and the level of apoptosis are decreased (see Fig. 4). The inhibitor also leads to a decrease in caspase 3 activity (Fig. 5 A and B) and PARP cleavage (Fig. 5 C and D). It is interesting to speculate that the MEK inhibitor may have an as yet undefined effect on the cell cycle, thereby leading to the reversal of the G2/M block. Certainly, precedent for the flavopiridol compound family, to which the MEK inhibitor belongs, affecting cell cycle has been set (38–40). Alternatively, it may also be that a downstream MAPK from MEK has a direct effect on caspase activity. We have determined that this effect is not caused by the direct inactivation of the caspases by the inhibitor or that the GSNO is in some way altered upon contact with the MEK inhibitor, because the spectrum of the GSNO remains unchanged after incubation with the inhibitor (data not shown). A model for the action of PD 098059 on RAW and RES cells based on these observations is shown in Fig. 6.
Figure 6.
Potential effect of PD 908059 on apoptosis induced by NO in RAW and RES macrophages.
The regulation of caspases via the MAPK pathway is beginning to be defined, and several reports have recently linked the kinases to caspase regulation (41, 42). In either case, it certainly appears that the MEK inhibitor can compete with the apoptotic effect of exogenous NO. The exact mechanism whereby inhibition of the MEK pathway can block apoptosis induced via NO needs to be defined.
These findings demonstrate that adaptation to one type of apoptotic signal may render cells more susceptible to alternative apoptotic signals. The hypersensitive response of the RES cells may be directly caused by the selection process via LPS/IFN-γ or, alternatively, may be caused by the process of adaptation to apoptosis. The RES cell population is a valuable tool to examine apoptosis in response to NO. It is convenient that although susceptible to exogenous NO-mediated apoptosis, the RES cells do not undergo apoptosis to signals from iNOS, the endogenous source for NO. These cells should allow us to more closely define the relationship between NO and apoptosis.
Indeed, these cells may suggest a unique pathway(s) that may be exploited via exogenous NO that would induce apoptosis in cells that would normally be resistant to apoptosis. Cells that normally avoid apoptosis may contribute to pathology, as is the proposal for the contribution of macrophages within the atheroma. By delineating a mechanism that will cause apoptosis in resistant cells, elimination of pathological cell populations may be possible. By exploiting this mechanism, targeted apoptosis may be a possible way to control unwanted cell populations. The refinement of this mechanism of targeting apoptosis is ongoing in our laboratory.
Acknowledgments
This work was supported by grants from the Cleveland Foundation (E.G.L.), American Heart Association (SDG #973024ON) (T.S.M.), and Deutsche Forschungsgemeinschaft (Mo779/1–1), Germany (S.M.).
ABBREVIATIONS
- PARP
poly(ADP-ribose) polymerase
- GSNO
S-nitrosoglutathione
- LPS
lipopolysaccharide
- IFN-γ
interferon γ
- DEVD-AFC
N-acetyl-Asp-Glu-Val-Asp-7-amino-4-trifluoromethyl coumarin
- iNOS
inducible NO synthase
- MEK
mitogen-activated protein kinase kinase
- DEVD-CHO
N-acetyl-Asp-Glu-Val-Asp-aldehyde
- SAPK
stress-activated protein kinase
- MAPK
mitogen-activated protein kinase
References
- 1.Vaux D L, Haecker G, Strasser A. Cell. 1994;76:777–779. doi: 10.1016/0092-8674(94)90350-6. [DOI] [PubMed] [Google Scholar]
- 2.Raff M C. Nature (London) 1992;356:397–400. doi: 10.1038/356397a0. [DOI] [PubMed] [Google Scholar]
- 3.Wyllie A H, Kerr J F, Currie A R. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 4.Leist M, Nicotera P. Biochem Biophys Res Commun. 1997;236:1–9. doi: 10.1006/bbrc.1997.6890. [DOI] [PubMed] [Google Scholar]
- 5.Golstein P. Science. 1997;275:1081–1082. doi: 10.1126/science.275.5303.1081. [DOI] [PubMed] [Google Scholar]
- 6.Wyllie A H. Nature (London) 1994;369:272–273. doi: 10.1038/369272a0. [DOI] [PubMed] [Google Scholar]
- 7.Orth K, O’Rourke K, Salvesen G S, Dixit V M. J Biol Chem. 1996;271:20977–20980. doi: 10.1074/jbc.271.35.20977. [DOI] [PubMed] [Google Scholar]
- 8.Alnemri E, Livingston D J, Nicholson D W, Salvesen G, Thornberry N A, Wong W W, Yuan J. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. (lett.). [DOI] [PubMed] [Google Scholar]
- 9.Takahashi A, Earnshaw W C. Curr Opin Genet Dev. 1996;6:50–55. doi: 10.1016/s0959-437x(96)90010-6. [DOI] [PubMed] [Google Scholar]
- 10.Zhivotovsky B, Burgess D H, Vanags D M, Orrenius S. Biochem Biophys Res Commun. 1997;230:481–488. doi: 10.1006/bbrc.1996.6016. [DOI] [PubMed] [Google Scholar]
- 11.Porter A G, Ng P, Janicke R U. BioEssays. 1997;19:501–507. doi: 10.1002/bies.950190609. [DOI] [PubMed] [Google Scholar]
- 12.Mohr S, Zech B, Lapetina E G, Brüne B. Biochem Biophys Res Commun. 1997;238:387–391. doi: 10.1006/bbrc.1997.7304. [DOI] [PubMed] [Google Scholar]
- 13.Nathan C. FASEB J. 1992;6:3051–3064. [PubMed] [Google Scholar]
- 14.Nathan C, Xie Q W. J Biol Chem. 1994;269:13725–13728. [PubMed] [Google Scholar]
- 15.Walter U. Rev Phys Biochem Pharmacol. 1989;113:41–88. doi: 10.1007/BFb0032675. [DOI] [PubMed] [Google Scholar]
- 16.Wink D A, Hanbauer I, Grisham M B, Laval F, Nims R W, Laval J, Cook J, Pacelli R, Liebmann J, Krishna M, et al. Curr Top Cell Regul. 1996;34:159–187. doi: 10.1016/s0070-2137(96)80006-9. [DOI] [PubMed] [Google Scholar]
- 17.Mohr S, Stamler J S, Brüne B. FEBS Lett. 1994;348:223–227. doi: 10.1016/0014-5793(94)00596-6. [DOI] [PubMed] [Google Scholar]
- 18.Mohr S, Stamler J S, Brüne B. J Biol Chem. 1996;271:4209–4214. doi: 10.1074/jbc.271.8.4209. [DOI] [PubMed] [Google Scholar]
- 19.Stamler J S, Simon D I, Osborne J A, Mullins M E, Jaraki O, Michel T. Proc Natl Acad Sci USA. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamler J S. Cell. 1994;78:931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 21.Hirvonen M-R, Brüne B, Lapetina E G. Biochem J. 1996;315:845–849. doi: 10.1042/bj3150845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brüne B, Gotz C, Meβmer U K, Sandau K, Hirvonen M-R, Lapetina E G. J Biol Chem. 1997;272:7253–7258. doi: 10.1074/jbc.272.11.7253. [DOI] [PubMed] [Google Scholar]
- 23.Messmer U K, Brüne B. Biochem J. 1996;319:299–305. doi: 10.1042/bj3190299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messmer U K, Reed U K, Brüne B. J Biol Chem. 1996;271:20192–20197. doi: 10.1074/jbc.271.33.20192. [DOI] [PubMed] [Google Scholar]
- 25.Albina J E, Cui S, Mateo R B, Reichner J S. J Immunol. 1993;150:5080–5085. [PubMed] [Google Scholar]
- 26.Sarih M, Souvannavong V, Adam A. Biochem Biophys Res Commun. 1993;191:503–508. doi: 10.1006/bbrc.1993.1246. [DOI] [PubMed] [Google Scholar]
- 27.Endres M, Wang Z Q, Namura S, Waeber C, Moskowitz M A. J Cereb Blood Flow Metab. 1997;17:1143–1151. doi: 10.1097/00004647-199711000-00002. [DOI] [PubMed] [Google Scholar]
- 28.Tzeng E, Kim Y M, Pitt B R, Lizonova A, Kovesdi I, Billiar T R. Surgery. 1997;122:255–263. doi: 10.1016/s0039-6060(97)90016-7. [DOI] [PubMed] [Google Scholar]
- 29.Mannick J B, Miao X Q, Stamler J S. J Biol Chem. 1997;272:24125–24128. doi: 10.1074/jbc.272.39.24125. [DOI] [PubMed] [Google Scholar]
- 30.Li J, Billiar T R, Talanian R V, Kim Y M. Biochem Biophys Res Commun. 1997;240:419–424. doi: 10.1006/bbrc.1997.7672. [DOI] [PubMed] [Google Scholar]
- 31.Kim Y M, Talanian R V, Billiar T R. J Biol Chem. 1997;272:31138–31148. doi: 10.1074/jbc.272.49.31138. [DOI] [PubMed] [Google Scholar]
- 32.Brüne B, Golkel C, von Knethen A. Biochem Biophys Res Comm. 1996;229:396–401. doi: 10.1006/bbrc.1996.1816. [DOI] [PubMed] [Google Scholar]
- 33.Hampton M B, Orrenius S. FEBS Lett. 1997;414:552–556. doi: 10.1016/s0014-5793(97)01068-5. [DOI] [PubMed] [Google Scholar]
- 34.Sarkar R, Gordon D, Stanley J C, Webb R C. Am J Physiol. 1997;272:H1810–H1818. doi: 10.1152/ajpheart.1997.272.4.H1810. [DOI] [PubMed] [Google Scholar]
- 35.Sciorati C, Nistico G, Meldolesi J, Clementi E. Br J Pharmacol. 1997;122:687–697. doi: 10.1038/sj.bjp.0701413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gansauge S, Gansauge F, Nussler A K, Rau B, Poch B, Schoenberg M H, Beger H G. FEBS Lett. 1997;410:160–164. doi: 10.1016/s0014-5793(97)00544-9. [DOI] [PubMed] [Google Scholar]
- 37.Takagi K, Isobe Y, Yasukawa K, Okouchi E, Suketa Y. FEBS Lett. 1994;340:159–162. doi: 10.1016/0014-5793(94)80128-2. [DOI] [PubMed] [Google Scholar]
- 38.Alessi D R, Cuenda A, Cohen P, Dudley D T, Saltiel A R. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 39.Carlson B A, Dubay M M, Sausville E A, Brizuela L, Worland P J. Cancer Res. 1996;56:2973–2978. [PubMed] [Google Scholar]
- 40.Konig A, Schwartz G K, Mohammad R M, Al-Katib A, Gabrilove J L. Blood. 1997;90:4307–4312. [PubMed] [Google Scholar]
- 41.Toyoshima F, Moriguchi T, Nishida E. J Cell Biol. 1997;139:1005–1015. doi: 10.1083/jcb.139.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang S, Jiang Y, Li Z, Nishida E, Mathias P, Lin S, Ulevitch R J, Nemerow G R, Han J. Immunity. 1997;6:739–749. doi: 10.1016/s1074-7613(00)80449-5. [DOI] [PubMed] [Google Scholar]