Abstract
We disrupted the fibroblast growth factor (FGF) receptor 2 (FGFR2) gene by introducing a neo cassette into the IIIc ligand binding exon and by deleting a genomic DNA fragment encoding its transmembrane domain and part of its kinase I domain. A recessive embryonic lethal mutation was obtained. Preimplantation development was normal until the blastocyst stage. Homozygous mutant embryos died a few hours after implantation at a random position in the uterine crypt, with collapsed yolk cavity. Mutant blastocysts hatched, adhered, and formed a layer of trophoblast giant cells in vitro, but after prolonged culture, the growth of the inner cell mass stopped, no visceral endoderm formed, and finally the egg cylinder disintegrated. It follows that FGFR2 is required for early postimplantation development between implantation and the formation of the egg cylinder. We suggest that FGFR2 contributes to the outgrowth, differentiation, and maintenance of the inner cell mass and raise the possibility that this activity is mediated by FGF4 signals transmitted by FGFR2. The role of early FGF signaling in pregastrulation development as a possible adaptation to mammalian (amniote) embryogenesis is discussed.
The fibroblast growth factor (FGF) system consists of four receptors (FGFR1–4) and 15 growth factors (FGF1–15). Signaling through FGFRs results in proliferation and differentiation of cultured cells. In vivo they contribute to multiple events of embryogenesis, from late gastrulation to various aspects of organogenesis (1–8). In the adult they have been implicated in angiogenesis, wound healing, tumorigenesis (9–11), and congenital dominant craniofacial and limb defects (for review see ref. 12).
The earliest acting member of the FGF system is FGF4. Its targeted loss-of-function phenotype results in embryonic death soon after implantation (1). FGF4 is first expressed during cleavage (13); later it is active in the blastocyst, in the egg cylinder, and then in the primitive streak (14). Between the blastocyst and primitive streak stages, implantation takes place, the first lineage decisions are made, the main body axes are laid down, and precursors of the extraembryonic tissues are formed (15). The latter establishes the typical mammalian fetal–maternal relationship. Much has been learned about the molecular mechanism of gastrulation, which is shared by all vertebrates. Less information is, however, available on early mammalian development and its aspects specific for amniote embryogenesis.
This investigation set out to study the nature of the receptor or receptors that transmit early FGF signals. In situ hybridization patterns suggested that FGFR2 may be one of the earliest-acting FGF receptors (16). Early expression has been detected with the highly sensitive reverse transcription (RT)-PCR assay also for FGFR3 and FGFR4 (13). More recent gene targeting experiments revealed loss-of-function phenotypes of three FGF receptors. FGFR1 is required during gastrulation for morphogenetic movements through the primitive streak (17). FGFR3 was shown to be a negative regulator of long bone development (18, 19), whereas targeted FGFR4 mutants showed no phenotype (C.-X. Deng, personal communication). We therefore assumed that FGFR2 may be a good candidate for the transduction of early FGF signals. Here we report gene targeting experiments, which suggest that FGFR2 is required for early postimplantation embryogenesis. Our data suggest that FGFR2 contributes to visceral endoderm differentiation and to the growth and maintenance of the inner cell mass (ICM).
MATERIALS AND METHODS
Animals.
Random-bred MF1 (Harlan Laboratories, Ein Karem, Jerusalem) and inbred 129/SvPas mice were used.
Gene targeting was performed by aggregation using R1 embryonic stem (ES) cells (20). The construct was prepared from a 129SvJ genomic λ phage library.
Embryo Culture.
Two- to eight-cell embryos were grown in drops of M16 medium under liquid paraffin (BDH). Blastocysts were grown in ES cell medium without lymphocyte inhibiting factor (LIF) (20) on gelatin-coated tissue culture plates. After 2 days the positions of the blastocysts that adhered were registered. They were individually photographed and harvested with Eppendorf tips for PCR.
PCR.
Embryos grown in vitro were placed into 200 μl of lysis buffer for DNA extraction (21). Two primer pairs were used. One pair, 5′-AGAGGCTATTCGGCTATGACTG-3′ and 5′-TTCGTCCAGATCATCCTGATC-3′, recognized the neo gene, whereas the other, taken from the two ends of the IIIc exon, 5′-GCCGCCGGTGTTAACACC-3′ and 5′-CTGGCAGAACTGTCAACCA-3′ recognized the wild-type receptor and was absent from homozygous mutants. The IIIc signal was detected by Southern blot hybridization.
Immunochemistry.
The visceral endoderm of cultured blastocysts was detected with biotin-conjugated Sophora japonica lectin (22) and visualized with ExtrAvidin Cy3 (Sigma). A sheep antibody against the complete ectodomain of human FGFR2 IIIc (Binding Site, Birmingham, U.K.) was also used. The antibody was characterized by its reactivity with mouse or human FGFR2 expressed in BAF cells, but not with FGFR1, FGFR3, or FGFR4, or with control BAF cells assayed by Western blotting or immunofluorescence. To detect FGFR2 in the cell membrane of cultured blastocysts, staining was performed without fixation, in M2 medium containing 0.1% BSA and 0.02% sodium azide. For confocal microscopy, blastocysts were stained in suspension after fixation with methanol, according to Larue et al. (23).
Histology and in Situ Hybridization.
Implantation sites between 4.25 and 5.5 days post coitus (d.p.c.) were detected by an intravenously injected vital dye, Chicago sky blue. Uterus fragments with implantation sites were fixed and embedded in paraffin, and in situ hybridization with FGFR2 and FGFR1 probes was performed as previously (16). Embryos (4.25–6.5 d.p.c.) were serially sectioned.
RESULTS
Targeted Disruption of FGFR2.
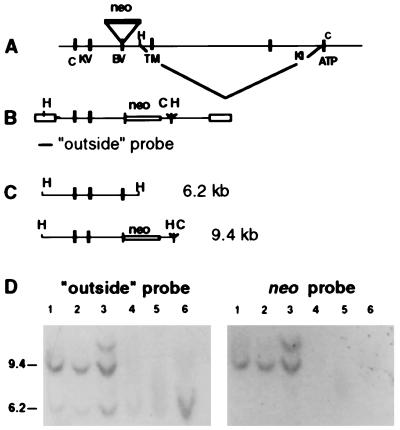
A neo cassette, controlled by the PGK-1 (3-phosphoglycerate kinase) promoter, was inserted in reverse transcriptional orientation into the EcoRV site of exon 9, encoding the IIIc Ig-like loop of the ligand binding domain (24). In addition, we deleted a 10- to 13-kb-long genomic DNA fragment that connects a HindIII site upstream of transmembrane exon 10 with a ClaI site in exon 12, encoding the ATP binding site of the first kinase domain (Fig. 1A–C). Our construct had the potential to create multiple defects. It abolished the IIIc transcriptional alternative. By reversing the transcriptional orientation of neo, multiple translational stop signals were created and the deletion abrogated the receptor’s membrane anchorage and enzymatic activity.
Figure 1.
Targeted disruption of FGFR2. (A) Genomic map and its manipulation. Above the line: H, HindIII; C, ClaI. Under the line: C, KV, and BV, exons 7, 8, and 9 (IIIc loop); TM, exon 10 (transmembrane); K1, kinase domain 1; ATP, exon 12 encoding the ATP binding site; neo, neomycin resistance gene. (B) Map of the recombinant, flanking sequences blocked, abbreviations as in A. (C) Wild-type (6.2-kb) and recombinant (9.4-kb) HindIII fragments. (D) Southern analysis of recombinant ES cells. Clones shown in lanes 1 and 2 were used for aggregation. The recombinant allele was analyzed also by a 3′ probe demonstrating the large deletion and the absence of the EcoRV site of exon 9. Specific probes showed the absence of the transmembrane domain.
The construct was electroporated into R1 ES cells (20). Neomycin-resistant homologous recombinant clones (15%) were isolated on the basis of Southern blot analysis (Fig. 1D). Two homologous recombinant ES cell lines were aggregated with eight-cell embryos (20), and after their transmission to the germ line, two noninbred MF1-based mouse strains were established. Heterozygotes of both strains were normal and fertile. Inbred mice on 129/SvPas background were also derived, but all strains displayed similar phenotypes.
The FGFR2 Mutant Dies Soon After Implantation.
Mating FGFR2 heterozygotes did not produce live homozygous offspring, and no homozygous mutant embryos were found between 8.5 and 18.5 days of gestation (Table 1). Moreover, very few decidua contained dead embryos, suggesting that the mutant dies before a visible decidual reaction. To investigate the earliest postimplantation stages, vital stained implantation sites (4.5, 5.0, 5.5, and 6.0 d.p.c) were subjected to histological analysis. All implantation sites were sectioned serially, and the ones with embryos visible at low power were selected for further observation. A number of “empty” decidua were, however, investigated at high power without finding distinguishable remnants of the embryo. Twenty-two 5.5 and 6.0 d.p.c. implantation sites contained no defective embryos. Three of 21 were abnormal at day 5.0 of gestation. At day 4.5, in contrast, a quarter (10 of 38) displayed characteristic defects (Fig. 2). This percentage was consistent with a single recessive lethal allele. It appears that increased capillary permeability, which indicates the first step of implantation, did not trigger full decidualization by the presumptive mutant embryo. Because empty implantation sites did not develop into fully decidualized ampullae, it appears that the uterine reaction induced by the mutant trophectoderm was incomplete.
Table 1.
Genotypes of progeny from FGFR2+/− crosses
| Age | No. with genotype
|
|||
|---|---|---|---|---|
| +/− | +/+ | −/− | Total | |
| Weanling | 89 | 40 | — | 129 |
| 19.5 d.p.c. | 10 | 3 | — | 13 |
| 10.5 d.p.c. | 25 | 8 | — | 33 |
| 8.5 d.p.c. | 17 | 4 | — | 21 |
| Cultured blastocysts | 34 | 17 | 12 | 63 |
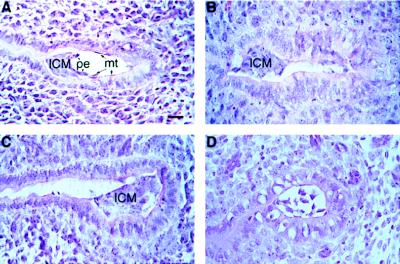
Figure 2.
Histological analysis of 4.5 d.p.c. (FGFR2+/−)F2 embryos. (A) Control. (B–D) Presumptive mutants. The mesometrium is toward the left side of the figures. pe, Primitive endoderm; mt, mural trophectoderm. (Bar = 25 μm.)
Defective embryos could be distinguished by their random position with respect to the mesometrial-antimesometrial extent of the uterine crypt and by the absence of the former blastocele. They attached to the uterine epithelium but, instead of the canonical mesometrial position of the ICM (Fig. 2A), the defective embryo attached to the side (Fig. 2B) or to the antimesometrial end of the crypt (Fig. 2C), and in some embryos the ICM entirely disaggregated (Fig. 2D). Most 4.5-d.p.c. normal embryos displayed a row of darkly stained presumptive primitive endoderm cells facing the yolk cavity (Fig. 2A). This cell layer was not seen in the abnormal embryo.
FGFR2 Is Required for ICM Differentiation in Vitro.
First, two- to four-cell embryos deriving from (FGFR2+/−)F2 or as control (FGFR2+/− × +/+) backcross matings were cultured until the blastocyst stage. No defects or differences between the two groups were observed, suggesting that the mutant can form normal blastocysts. Next, to simulate implantation, 3.5-d.p.c. blastocysts were explanted on gelatin-coated tissue culture plates. During the first 2 to 3 days, both F2 and backcross blastocysts hatched from the zona, adhered, and developed a giant trophoblast layer around the incipient egg cylinder (compare Fig. 3B–D with E and G). Soon afterward, however, the ICM of the mutant stopped expanding and disintegrated without developing a discernible visceral endoderm layer (Fig. 3 B–D). In all mutants the giant trophoblast layer survived the disintegration of the ICM (Fig. 3 C and D) and was alive at the end of culture (day 5 to 7).
Figure 3.
Mutant defects in cultured blastocysts; FGFR2 expression in preimplantation blastocysts. (A–D) Seven-day cultures. (A) Heterozygote. (B–D) Homozygous mutant. (E–H) Four-day cultures, Sophora japonica lectin staining. (E and F) Heterozygote. (G and H) Homozygous mutant. (E and G) Phase contrast. (F and H) Fluorescence. (I) Blastocyst (3.5 d.p.c.) anti-FGFR2 fluorescent staining, confocal microscopy. (Bars: 250 μm.)
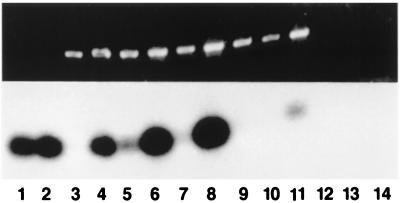
The genotype of normal and defective blastocysts was analyzed by PCR. The above phenotype was in full concordance with the mutant genotype (Fig. 4), with allele ratios consistent with a single recessive allele (bottom line, Table 1). Hence both our in vivo and in vitro observations suggest that FGFR2 is required for early postimplantation embryogenesis.
Figure 4.
PCR analysis of individual cultured blastocysts. (Upper) Ethidium bromide staining. (Lower) Southern blot hybridization with FGFR2 probe. Lanes 1 and 2, homozygous wild type; lanes 4, 5, 6, 8, and 11, heterozygotes; lanes 3, 7, 9, and 10, homozygous mutant; lanes 12 and 13, blank control; lane 14, water. This is an analysis of the experiment visualized in part in Fig. 3 A–D.
The ICM differentiates into the primitive endoderm and its derivative the visceral endoderm, which surrounds the egg cylinder (15). We investigated the contribution of FGFR2 to endoderm differentiation by the visceral endoderm-specific Sophora japonica lectin (22) at day 4 of culture, when little visible defect could be observed (Fig. 3 E–H). Biotin-avidin cytochemistry, with individual PCR genotyping, revealed much less signal surrounding mutant (Fig. 3 G and H) than heterozygous wild-type egg cylinders (Fig. 3 E and F), indicating that FGFR2 is required for normal ICM differentiation.
Localization of FGFR2 in the Early Embryo.
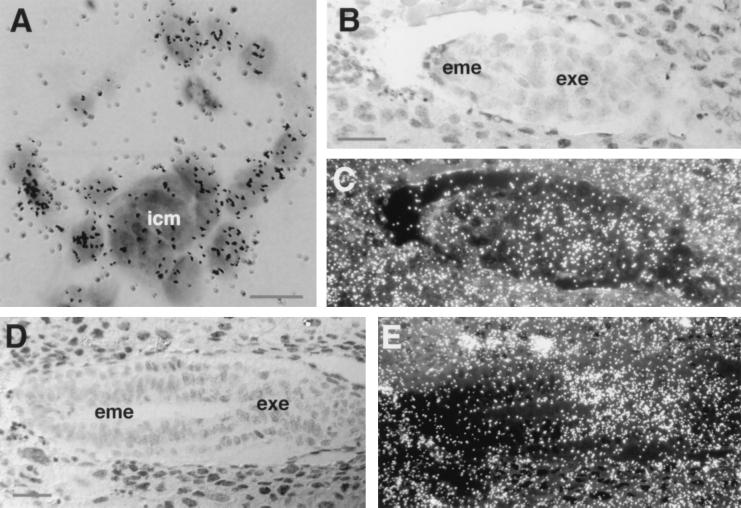
We previously described the localized expression of FGFR2 from 6.5 d.p.c. throughout embryogenesis (16). Because the present data suggested that FGFR2 is required already at 4.5 d.p.c., we decided to explore the relevant period. Immunochemistry and in situ hybridization were used to this end. Confocal microscopy with an antibody specific to the ectodomain of FGFR2 demonstrated that FGFR2 is present on the membrane and in the cytoplasm of most cells of preimplantation blastocysts (Fig. 3I). In situ hybridization gave similar results on free (Fig. 5A) and freshly implanted (not shown) blastocysts, with the difference that transcription was more pronounced in the trophectoderm than in the ICM. This trend increased as egg cylinder development progressed. At 5.5 and 6.5 d.p.c., FGFR2 transcripts localized to the trophectoderm derived extraembryonic ectoderm (Fig. 5 B–E), and by these stages no expression was detectable in the visceral endoderm. We conclude that FGFR2 is expressed in most cells of the early blastocyst. The pattern becomes more specialized during the egg cylinder stage, when it concentrates to the trophectoderm and its derivative the extraembryonic ectoderm.
Figure 5.
Localization of FGFR2 transcripts in the early embryo by in situ hybridization. (A) Blastocyst 3.5 d.p.c. (B and D) Bright field. (C and E) Dark field. (B and C) Embryo 5.5 d.p.c. (D and E) Embryo 6.5 d.p.c. (Bars: A = 20 μm; B–E = 200 μm.)
DISCUSSION
We demonstrated here that FGFR2 is required for early postimplantation development. FGFR2 has two splice variants with different binding specificities (25). The IIIc variant binds and transmits mitogenic signals from FGF1, -2, -4, -6, -8b, and -9, whereas the IIIb (KGFR) variant recognizes FGF1, -3, and -7 (26). Hence FGF4 is a potential ligand for FGFR2-IIIc. FGF4 has the earliest known expression pattern among FGFs. It is present in the two-cell embryo, as seen by PCR (13), and its transcripts are detectable by in situ hybridization in the ICM of mature blastocysts. In late egg cylinder embryos, after a more diffuse early expression pattern, FGF4 transcripts concentrate to the primitive streak (14). Comparing the localized transcription of FGF4 and FGFR2 reveals the close proximity of their expression in preimplantation and early postimplantation embryos (16, 27). Close functional connection between FGF4 and FGFR2 was indicated by the similarity of their targeted phenotypes, whereas loss of function of other FGFRs affected later events (2, 3, 18, 19). It follows that the relationship between FGF4 and FGFR2 fulfills three criteria indicating functional interaction: they posses mutual binding affinity, they are coexpressed, and their loss-of-function phenotypes are similar. We therefore think that FGF4 and FGFR2 interact during a period that falls between implantation and the outgrowth of the egg cylinder. Additional mutagenesis and expression studies of new members of the FGF family, and as-yet-uninvestigated FGFR transcriptional variants, will test this hypothesis.
In our FGFR2 mutation, translational elongation, binding specificity, membrane anchorage, and enzymatic activity were all abrogated. The group of C. Deng recently reported another targeted FGFR2 mutation (28). They removed exons 7, 8, and 9, which together encode the third Ig-like loop of the ligand binding domain, whereas the transmembrane and enzymatic domains, as well as the first two Ig-like loops of FGFR2, remained intact. The result was midgestation lethality with placental defects, coupled with abrogated limb outgrowth and down-regulated FGF8 and FGF10 expression. Comparing the two targeted genotypes to their respective phenotypes suggests that the mutation reported here may represent the null phenotype of FGFR2.
Our homozygous mutant had the potential to synthesize a soluble fragment consisting of the first and second Ig-like domains. Such a fragment was not detectable in the cell membrane (unpublished data). Being a part of the ligand binding domain, it nevertheless could compete for the ligand and create dominant-negative effects. This effect, however, was not observed, since the heterozygotes were normal and fertile.
Despite the absence of FGFR2 activity, the ICM and trophectoderm of the blastocyst appeared to be normal, but soon after implantation, or somewhat later in its in vitro equivalent, the ICM ceased to thrive and the embryo died. Experiments with a visceral endoderm-specific lectin revealed defective endoderm differentiation. This observation is supported by recent experiments in our laboratory. Y.C., I. Rubin, J.K.H., and P.L. (unpublished results) showed that abrogated FGF signaling in embryoid bodies expressing truncated dominant-negative FGFR2 cDNA inhibits endoderm differentiation. Abrogated endoderm development was connected to the down-regulation of visceral endoderm-specific genes, including α-fetoprotein, GATA-4, HNF-4, and Evx-1 (29, 30). This connection supports our suggestion that FGF signaling is required to induce endoderm differentiation in the ICM. The primitive endoderm and its visceral and parietal endoderm derivatives are components of the yolk sac. Hence FGF signaling appears to be involved in the development of this typical extraembryonic structure.
Differentiation of the ICM into primitive endoderm represents the second cell lineage decision of the mammalian embryo, whereas the first lineage decision involves the trophectoderm (15). Our data suggest that FGFR2 may be required for certain trophectoderm functions. Mutant blastocysts induce increased capillary permeability, but no extensive uterine reaction. Hence in the absence of FGFR2 trophectoderm-induced decidualization (31) is incomplete. Trophectoderm defects and the resulting abrogation of fetal–maternal interactions may be responsible for the early in vivo death and also for the random position of the embryo in the uterine crypt. The latter may indicate a role for FGF signaling in early embryonic polarity.
FGFR2 joins a group of genes that display early lethal loss-of-function phenotypes. The earliest is E-cadherin, a cell adhesion molecule required for trophoblast development (23). FGF4 (1), β1 integrin (32, 33), and Evx-1 (34) display phenotypes similar to that described in the present report for FGFR2. FGF4 is a potential ligand of FGFR2. Evx-1 encodes a homeobox protein. Its expression is connected with FGF signaling both in limb development (35) and in the differentiation of embryoid bodies (Y.C., I. Rubin, J.K.H., and P.L., unpublished results). Integrins mediate cell-to-cell and cell-to-matrix adhesion, and they are associated with FGF receptors and other receptor tyrosine kinases (36).
The role of FGF signaling in amphibian development has been investigated in great detail. FGF was among the first mesoderm inducers to be described (37, 38). Dominant-negative loss of FGFR function abrogated posterior lateral mesoderm differentiation in the late Xenopus gastrula (39). In contrast, targeted disruption of FGF4 and FGFR2 in the mouse resulted a considerably earlier phenotype. It is possible that FGFR2, and the gene activities associated with it, represent an evolutionary adaptation to mammalian or amniote embryogenesis.
Acknowledgments
We thank Dr. C. Deng (National Institutes of Health, Bethesda, MD) for sharing his data before publication. The help of Dr. A. Kniszinski and Dr. T. Burakova in the morula aggregation experiments, of Miss M. Fomin in histology, and of Mr. N. Tutka in animal breeding is gratefully acknowledged. This study was supported by The Infrastructure Laboratory for Gene Targeting, established by the Ministry of Science, and by a Center of Excellence of the Israel Science Fund.
ABBREVIATIONS
- FGF
fibroblast growth factor
- FGFR
FGF receptor
- ICM
inner cell mass
- ES cell
embryonic stem cell
- d.p.c.
days post coitus
References
- 1.Feldman B, Poueymirou W, Papaioannou V E, DeChiare T M, Goldfarb M. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- 2.Yamaguchi T P, Harpal K, Henkemeyer M, Rossant J. Genes Dev. 1994;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 3.Deng C-X, Wynshaw-Boris A, Shen M M, Daugherty C, Ornitz D M, Leder P. Genes Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 4.Mansour S L, Goddard J M, Capecchi M R. Development (Cambridge, UK) 1993;117:13–28. doi: 10.1242/dev.117.1.13. [DOI] [PubMed] [Google Scholar]
- 5.Cohn M J, Izpisua-Belmonte J C, Abud H, Heath J K, Tickle C. Cell. 1995;80:739–746. doi: 10.1016/0092-8674(95)90352-6. [DOI] [PubMed] [Google Scholar]
- 6.Post M, Souza P, Liu J, Tseu I, Wang J, Kuliszewski M, Tanswell A K. Development. 1996;122:3107–3115. doi: 10.1242/dev.122.10.3107. [DOI] [PubMed] [Google Scholar]
- 7.Neubuser A, Peters H, Balling R, Martin G R. Cell. 1997;90:247–255. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- 8.Hebert J M, Rosenquist T, Gotz J, Martin G M. Cell. 1994;78:1017–1025. doi: 10.1016/0092-8674(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 9.Flamme I, Risau W. Development (Cambridge, UK) 1992;116:435–439. doi: 10.1242/dev.116.2.435. [DOI] [PubMed] [Google Scholar]
- 10.Werner S, Smola H, Liao X, Longaker M T, Krieg T, Hofschneider H P, Williams L T. Science. 1994;266:819–822. doi: 10.1126/science.7973639. [DOI] [PubMed] [Google Scholar]
- 11.Folkman J. Nat Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 12.Wilkie A O M, Morriss-Kay G M, Jones E Y, Heath J K. Curr Biol. 1995;5:500–507. doi: 10.1016/s0960-9822(95)00102-3. [DOI] [PubMed] [Google Scholar]
- 13.Rappolee D A, Basilico C, Patel Y, Werb Z. Development (Cambridge, UK) 1994;120:2259–2269. doi: 10.1242/dev.120.8.2259. [DOI] [PubMed] [Google Scholar]
- 14.Niswander L, Martin G R. Development (Cambridge, UK) 1992;114:755–768. doi: 10.1242/dev.114.3.755. [DOI] [PubMed] [Google Scholar]
- 15.Gardner R L. Int Rev Exp Pathol. 1983;24:63–133. [PubMed] [Google Scholar]
- 16.Orr-Urtreger A, Givol D, Yayon A, Yarden Y, Lonai P. Development (Cambridge, UK) 1991;113:1419–1434. doi: 10.1242/dev.113.4.1419. [DOI] [PubMed] [Google Scholar]
- 17.Ciruna B G, Schwartz L, Harpal K, Yamaguchi T P, Rossant J. Development (Cambridge, UK) 1997;124:2829–2841. doi: 10.1242/dev.124.14.2829. [DOI] [PubMed] [Google Scholar]
- 18.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 19.Colvin J S, Bohne B A, Harding G W, McEwen D G, Ornitz D M. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 20.Nagy A, Rossant J, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, New York: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 22.Sato M, Muramatsu T. Differentiation. 1985;29:29–38. doi: 10.1111/j.1432-0436.1985.tb00289.x. [DOI] [PubMed] [Google Scholar]
- 23.Larue L, Ohsugi M, Hirchenhain J, Kemler R. Proc Natl Acad Sci USA. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Givol D, Yayon A. FASEB J. 1992;6:3362–3369. [PubMed] [Google Scholar]
- 25.Miki T, Bottaro D P, Fleming T P, Smith C L, Burgess W H, Chan A M, Aaronson S A. Proc Natl Acad Sci USA. 1992;89:246–250. doi: 10.1073/pnas.89.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ornitz D M, Xu J, Colvin J S, McEwen D G, MacArthur C A, Coulier F, Gao G, Goldfarb M. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 27.Orr-Urtreger A, Bedford M T, Burakova T, Arman E, Zimmer Y, Yayon A, Givol D, Lonai P. Dev Biol. 1993;158:475–486. doi: 10.1006/dbio.1993.1205. [DOI] [PubMed] [Google Scholar]
- 28.Xu, X., Weinstein, M., Li, C., Naski, M., Cohen, R. I., Ornitz, D. M., Leder, P. & Deng, C. (1998) Development (Cambridge, U.K.), in press. [DOI] [PubMed]
- 29.Soudais C, Bielinska M, Heikinheimo M, MacArthur C A, Narita N, Saffitz J E, Simon M C, Leiden J M, Wilson D B. Development (Cambridge, UK) 1995;121:3877–3888. doi: 10.1242/dev.121.11.3877. [DOI] [PubMed] [Google Scholar]
- 30.Duncan S A, Nagy A, Chan W. Development (Cambridge, UK) 1997;124:279–287. doi: 10.1242/dev.124.2.279. [DOI] [PubMed] [Google Scholar]
- 31.Gardner R L, Johnson M H. J Embryol Exp Morphol. 1972;28:279–312. [PubMed] [Google Scholar]
- 32.Fassler R, Meyer M. Genes Dev. 1995;9:1896–1908. doi: 10.1101/gad.9.15.1896. [DOI] [PubMed] [Google Scholar]
- 33.Stephens L E, Sutherland A E, Klimansjaya I V, Andrieux A, Meneses J, Pedersen R A, Damsky C H. Genes Dev. 1995;9:1883–1895. doi: 10.1101/gad.9.15.1883. [DOI] [PubMed] [Google Scholar]
- 34.Spyropoulos D D, Capecchi M R. Genes Dev. 1994;8:1949–1961. doi: 10.1101/gad.8.16.1949. [DOI] [PubMed] [Google Scholar]
- 35.Niswander L, Martin G R. Development (Cambridge, UK) 1993;119:287–294. doi: 10.1242/dev.119.1.287. [DOI] [PubMed] [Google Scholar]
- 36.Miyamoto S, Teramoto H, Coso O A, Gutkind J S, Burbelo P D, Akiyama S K, Yamada K M. J Cell Biol. 1995;131:791–805. doi: 10.1083/jcb.131.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slack J M, Darlington B G, Heath J K, Godsave S F. Nature (London) 1987;326:197–200. doi: 10.1038/326197a0. [DOI] [PubMed] [Google Scholar]
- 38.Kimelman D, Kirschner M. Cell. 1987;51:869–877. doi: 10.1016/0092-8674(87)90110-3. [DOI] [PubMed] [Google Scholar]
- 39.Amaya E, Musci T J, Kirschner M W. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]