Abstract
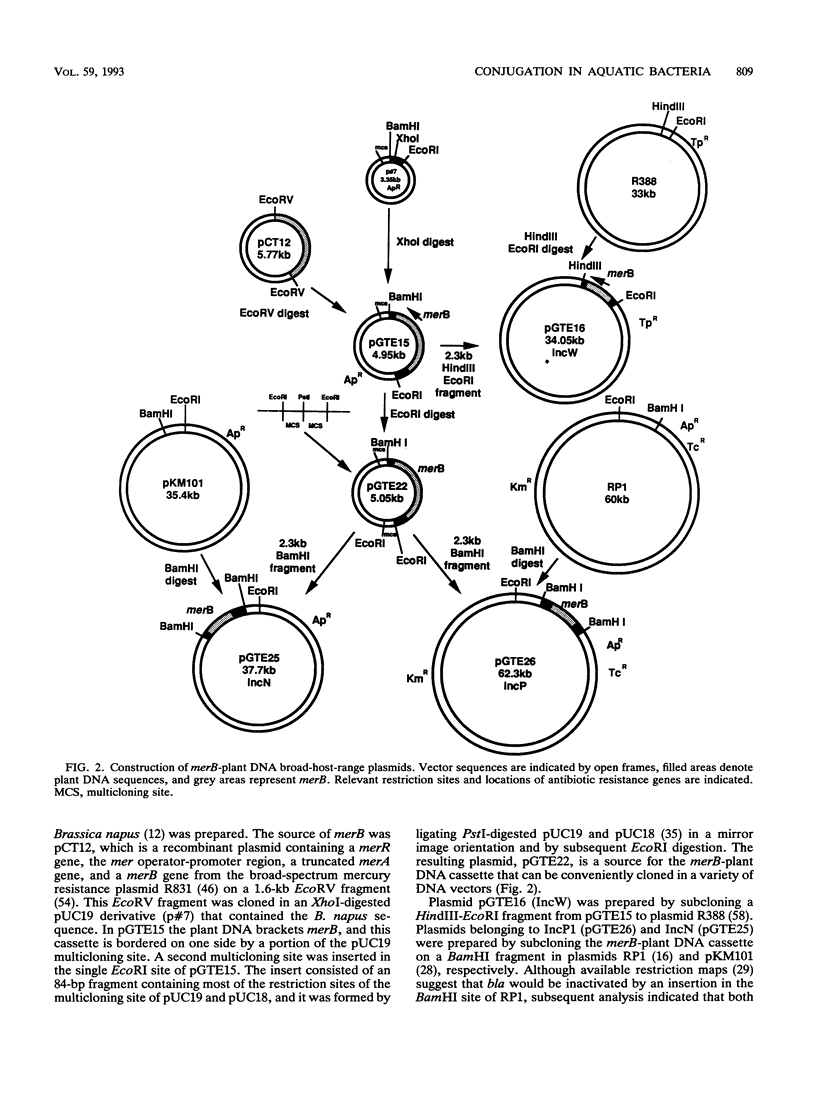
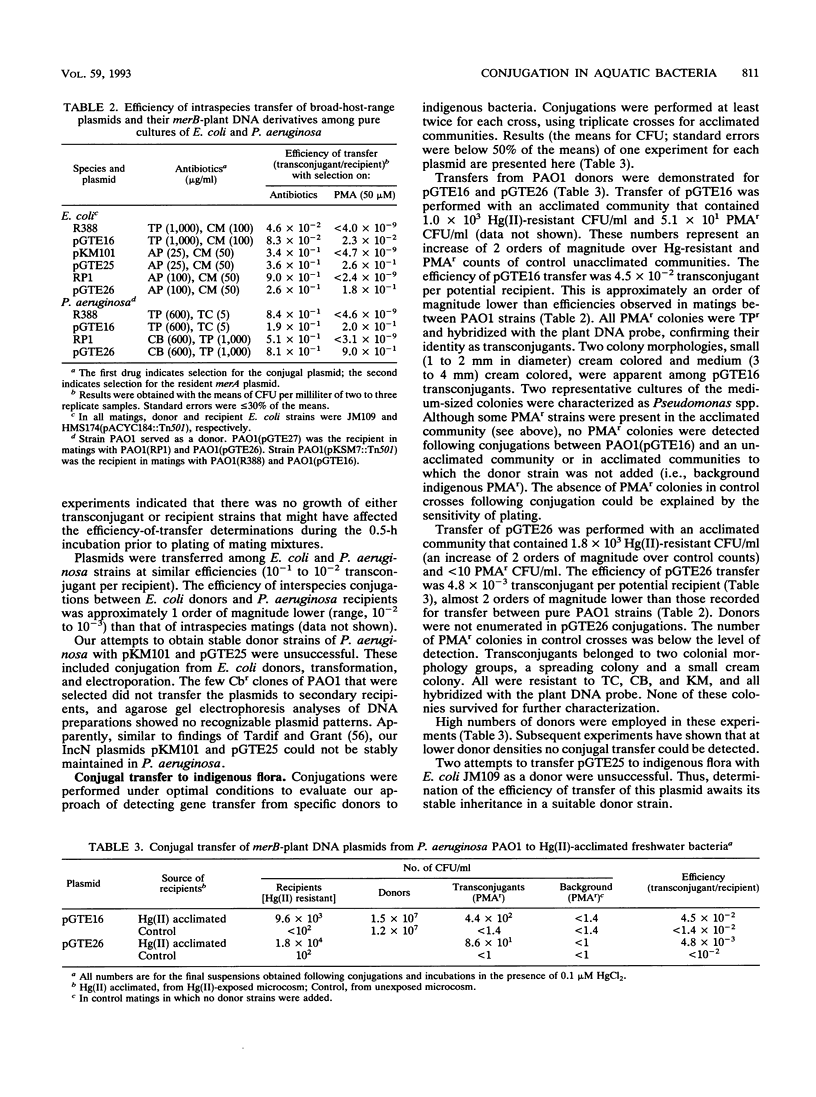
An experimental approach based on the assembly of genes of a catabolic pathway was used to detect transconjugants in aquatic communities. Resistance to phenylmercury acetate was established in transconjugants when wide-host-range conjugal plasmids containing merB, the gene encoding organomercurial lyase, were transferred to strains from aquatic communities that had been acclimated to inorganic mercury and thus enriched for populations containing merA, the gene encoding mercuric reductase (T. Barkay, Appl. Environ. Microbiol. 53:2725-2732, 1987). Conjugation was confirmed by using the plasmids' encoded antibiotic resistance patterns and by hybridization with a eukaryotic gene. Three merB-conjugal plasmids, belonging to incompatibility groups W (pGTE16), P1 (pGTE26), and N (pGTE25), were prepared. Transfers by filter matings of pGTE16 and pGTE26 from Pseudomonas aeruginosa PA01 to indigenous strains were at efficiencies of 4.5 × 10-2 and 4.8 × 10-3 transconjugant per potential recipient, respectively. These efficiencies were from 1 to 2 orders of magnitude below those observed for intraspecies matings with genetically marked recipients. The third plasmid, pGTE25, was not stably maintained in P. aeruginosa donors, and its transfer from Escherichia coli donors was below the level of detection. Characterized transconjugant strains were shown to be Pseudomonas spp. Potential applications of the described experimental approach in the creation of bacterial populations with new catabolic capabilities in hazardous waste sites and in the detection of transfer of recombinant DNA from engineered microorganisms to indigenous bacteria are discussed.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Timmis K. N. Host: vector systems for gene cloning in Pseudomonas. Curr Top Microbiol Immunol. 1982;96:47–67. doi: 10.1007/978-3-642-68315-2_4. [DOI] [PubMed] [Google Scholar]
- Barkay T. Adaptation of aquatic microbial communities to hg stress. Appl Environ Microbiol. 1987 Dec;53(12):2725–2732. doi: 10.1128/aem.53.12.2725-2732.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T. Adaptation of aquatic microbial communities to hg stress. Appl Environ Microbiol. 1987 Dec;53(12):2725–2732. doi: 10.1128/aem.53.12.2725-2732.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T., Gillman M., Liebert C. Genes encoding mercuric reductases from selected gram-negative aquatic bacteria have a low degree of homology with merA of transposon Tn501. Appl Environ Microbiol. 1990 Jun;56(6):1695–1701. doi: 10.1128/aem.56.6.1695-1701.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T., Liebert C., Gillman M. Environmental significance of the potential for mer(Tn21)-mediated reduction of Hg2+ to Hg0 in natural waters. Appl Environ Microbiol. 1989 May;55(5):1196–1202. doi: 10.1128/aem.55.5.1196-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T., Liebert C., Gillman M. Hybridization of DNA probes with whole-community genome for detection of genes that encode microbial responses to pollutants: mer genes and Hg2+ resistance. Appl Environ Microbiol. 1989 Jun;55(6):1574–1577. doi: 10.1128/aem.55.6.1574-1577.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T., Pritchard H. Adaptation of aquatic microbial communities to pollutant stress. Microbiol Sci. 1988 Jun;5(6):165–169. [PubMed] [Google Scholar]
- Bradley D. E. Morphological and serological relationships of conjugative pili. Plasmid. 1980 Sep;4(2):155–169. doi: 10.1016/0147-619x(80)90005-0. [DOI] [PubMed] [Google Scholar]
- Crosa J. H. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol Rev. 1989 Dec;53(4):517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch M. L., Tenbarge K. M., Simon A. E., Ferl R. cDNA clones for Brassica napus seed storage proteins: evidence from nucleotide sequence analysis that both subunits of napin are cleaved from a precursor polypeptide. J Mol Appl Genet. 1983;2(3):273–283. [PubMed] [Google Scholar]
- Fulthorpe R. R., Wyndham R. C. Involvement of a chlorobenzoate-catabolic transposon, Tn5271, in community adaptation to chlorobiphenyl, chloroaniline, and 2,4-dichlorophenoxyacetic acid in a freshwater ecosystem. Appl Environ Microbiol. 1992 Jan;58(1):314–325. doi: 10.1128/aem.58.1.314-325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D., You I. S., Chatterjee D. K., Chakrabarty A. M. Microbial degradation of halogenated compounds. Science. 1985 Apr 12;228(4696):135–142. doi: 10.1126/science.228.4696.135. [DOI] [PubMed] [Google Scholar]
- Grinsted J., Saunders J. R., Ingram L. C., Sykes R. B., Richmond M. H. Properties of a R factor which originated in Pseudomonas aeruginosa 1822. J Bacteriol. 1972 May;110(2):529–537. doi: 10.1128/jb.110.2.529-537.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Rekik M., Wasserfallen A., Bairoch A. Evolutionary relationships between catabolic pathways for aromatics: conservation of gene order and nucleotide sequences of catechol oxidation genes of pWW0 and NAH7 plasmids. Mol Gen Genet. 1987 Dec;210(2):241–247. doi: 10.1007/BF00325689. [DOI] [PubMed] [Google Scholar]
- Jain R. K., Burlage R. S., Sayler G. S. Methods for detecting recombinant DNA in the environment. Crit Rev Biotechnol. 1988;8(1):33–84. doi: 10.3109/07388558809150537. [DOI] [PubMed] [Google Scholar]
- Kellogg S. T., Chatterjee D. K., Chakrabarty A. M. Plasmid-assisted molecular breeding: new technique for enhanced biodegradation of persistent toxic chemicals. Science. 1981 Dec 4;214(4525):1133–1135. doi: 10.1126/science.7302584. [DOI] [PubMed] [Google Scholar]
- Kerr A., Manigault P., Tempé J. Transfer of virulence in vivo and in vitro in Agrobacterium. Nature. 1977 Feb 10;265(5594):560–561. doi: 10.1038/265560a0. [DOI] [PubMed] [Google Scholar]
- Kröckel L., Focht D. D. Construction of chlorobenzene-utilizing recombinants by progenitive manifestation of a rare event. Appl Environ Microbiol. 1987 Oct;53(10):2470–2475. doi: 10.1128/aem.53.10.2470-2475.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer P. J., Walker G. C. Restriction endonuclease cleavage map of pKM101: relationship to parental plasmid R46. Mol Gen Genet. 1981;182(2):268–272. doi: 10.1007/BF00269669. [DOI] [PubMed] [Google Scholar]
- Lanka E., Lurz R., Fürste J. P. Molecular cloning and mapping of SphI restriction fragments of plasmid RP4. Plasmid. 1983 Nov;10(3):303–307. doi: 10.1016/0147-619x(83)90047-1. [DOI] [PubMed] [Google Scholar]
- Lehrbach P. R., McGregor I., Ward J. M., Broda P. Molecular relationships between pseudomonas INC P-9 degradative plasmids TOL, NAH, and SAL. Plasmid. 1983 Sep;10(2):164–174. doi: 10.1016/0147-619x(83)90069-0. [DOI] [PubMed] [Google Scholar]
- Lehrbach P. R., Zeyer J., Reineke W., Knackmuss H. J., Timmis K. N. Enzyme recruitment in vitro: use of cloned genes to extend the range of haloaromatics degraded by Pseudomonas sp. strain B13. J Bacteriol. 1984 Jun;158(3):1025–1032. doi: 10.1128/jb.158.3.1025-1032.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984 Nov 16;226(4676):792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Miller R. V., Ku C. M. Characterization of Pseudomonas aeruginosa mutants deficient in the establishment of lysogeny. J Bacteriol. 1978 Jun;134(3):875–883. doi: 10.1128/jb.134.3.875-883.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., Siak J. S., Gray R. H. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J Virol. 1974 Sep;14(3):689–699. doi: 10.1128/jvi.14.3.689-699.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichard Scott L., Paul John H. Detection of Gene Expression in Genetically Engineered Microorganisms and Natural Phytoplankton Populations in the Marine Environment by mRNA Analysis. Appl Environ Microbiol. 1991 Jun;57(6):1721–1727. doi: 10.1128/aem.57.6.1721-1727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J. L., Wasserfallen A., Rose K., Timmis K. N. Redesigning metabolic routes: manipulation of TOL plasmid pathway for catabolism of alkylbenzoates. Science. 1987 Jan 30;235(4788):593–596. doi: 10.1126/science.3468623. [DOI] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Hybrid pathway for chlorobenzoate metabolism in Pseudomonas sp. B13 derivatives. J Bacteriol. 1980 May;142(2):467–473. doi: 10.1128/jb.142.2.467-473.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. E., Felton J., Wright A. Insertion of DNA activates the cryptic bgl operon in E. coli K12. Nature. 1981 Oct 22;293(5834):625–629. doi: 10.1038/293625a0. [DOI] [PubMed] [Google Scholar]
- Rojo F., Pieper D. H., Engesser K. H., Knackmuss H. J., Timmis K. N. Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science. 1987 Dec 4;238(4832):1395–1398. doi: 10.1126/science.3479842. [DOI] [PubMed] [Google Scholar]
- Schottel J., Mandal A., Clark D., Silver S., Hedges R. W. Volatilisation of mercury and organomercurials determined by inducible R-factor systems in enteric bacteria. Nature. 1974 Sep 27;251(5473):335–337. doi: 10.1038/251335a0. [DOI] [PubMed] [Google Scholar]
- Silver S., Walderhaug M. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiol Rev. 1992 Mar;56(1):195–228. doi: 10.1128/mr.56.1.195-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit E., van Elsas J. D., van Veen J. A., de Vos W. M. Detection of Plasmid Transfer from Pseudomonas fluorescens to Indigenous Bacteria in Soil by Using Bacteriophage phiR2f for Donor Counterselection. Appl Environ Microbiol. 1991 Dec;57(12):3482–3488. doi: 10.1128/aem.57.12.3482-3488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain J. C., Pritchard P. H., Bourquin A. W. Effects of adaptation on biodegradation rates in sediment/water cores from estuarine and freshwater environments. Appl Environ Microbiol. 1980 Oct;40(4):726–734. doi: 10.1128/aem.40.4.726-734.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanisich V. A. The properties and host range of male-specific bacteriophages of Pseudomonas aeruginosa. J Gen Microbiol. 1974 Oct;84(2):332–342. doi: 10.1099/00221287-84-2-332. [DOI] [PubMed] [Google Scholar]
- Tardif G., Grant R. B. Transfer of IncN plasmids to Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1982 Jul;22(1):142–144. doi: 10.1128/aac.22.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Effects of Hg, CH(3)-Hg, and Temperature on the Expression of Mercury Resistance Genes in Environmental Bacteria. Appl Environ Microbiol. 1990 Nov;56(11):3266–3272. doi: 10.1128/aem.56.11.3266-3272.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. M., Grinsted J. Mapping of functions in the R-plasmid R388 by examination of deletion mutants generated in vitro. Gene. 1978 Apr;3(2):87–95. doi: 10.1016/0378-1119(78)90053-7. [DOI] [PubMed] [Google Scholar]



