Abstract
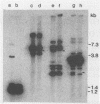
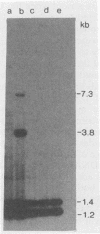
Three different methods of DNA isolation (organic deproteinization, potassium acetate deproteinization, and the use of cetyltrimethylammonium bromide) have been used to prepare DNA from Bacillus subtilis. Subsequent hybridization with an rDNA probe (DNA coding for rRNA) produces different patterns, which mirror those previously reported to indicate an rDNA deletion.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan E. J. Induction and cultivation of a stable L-form of Bacillus subtilis. J Appl Bacteriol. 1991 Apr;70(4):339–343. doi: 10.1111/j.1365-2672.1991.tb02946.x. [DOI] [PubMed] [Google Scholar]
- Barry T., Powell R., Gannon F. A general method to generate DNA probes for microorganisms. Biotechnology (N Y) 1990 Mar;8(3):233–236. doi: 10.1038/nbt0390-233. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Fry N. K., Warwick S., Saunders N. A., Embley T. M. The use of 16S ribosomal RNA analyses to investigate the phylogeny of the family Legionellaceae. J Gen Microbiol. 1991 May;137(5):1215–1222. doi: 10.1099/00221287-137-5-1215. [DOI] [PubMed] [Google Scholar]
- Loughney K., Lund E., Dahlberg J. E. Deletion of an rRNA gene set in Bacillus subtilis. J Bacteriol. 1983 Apr;154(1):529–532. doi: 10.1128/jb.154.1.529-532.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughney K., Lund E., Dahlberg J. E. tRNA genes are found between 16S and 23S rRNA genes in Bacillus subtilis. Nucleic Acids Res. 1982 Mar 11;10(5):1607–1624. doi: 10.1093/nar/10.5.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M. G., Thompson W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980 Oct 10;8(19):4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L. K., Dillon J. A. Molecular fingerprinting of isolates of the genus Peptostreptococcus using rRNA genes from Escherichia coli and P. anaerobius. J Gen Microbiol. 1991 Jun;137(6):1323–1331. doi: 10.1099/00221287-137-6-1323. [DOI] [PubMed] [Google Scholar]
- Ogasawara N., Moriya S., Yoshikawa H. Structure and organization of rRNA operons in the region of the replication origin of the Bacillus subtilis chromosome. Nucleic Acids Res. 1983 Sep 24;11(18):6301–6318. doi: 10.1093/nar/11.18.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen R. J. Chromosomal DNA fingerprinting--a new method of species and strain identification applicable to microbial pathogens. J Med Microbiol. 1989 Oct;30(2):89–99. doi: 10.1099/00222615-30-2-89. [DOI] [PubMed] [Google Scholar]
- Pickup R. W., Morgan J. A., Winstanley C., Saunders J. R. Implications for the release of genetically engineered organisms. Soc Appl Bacteriol Symp Ser. 1991;20:19S–30S. [PubMed] [Google Scholar]
- Rogall T., Flohr T., Böttger E. C. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J Gen Microbiol. 1990 Sep;136(9):1915–1920. doi: 10.1099/00221287-136-9-1915. [DOI] [PubMed] [Google Scholar]
- Rossau R., Duhamel M., Jannes G., Decourt J. L., Van Heuverswyn H. The development of specific rRNA-derived oligonucleotide probes for Haemophilus ducreyi, the causative agent of chancroid. J Gen Microbiol. 1991 Feb;137(2):277–285. doi: 10.1099/00221287-137-2-277. [DOI] [PubMed] [Google Scholar]
- Sandler N., Keynan A. Membrane binding and release of Bacillus subtilis DNA as a function of the cell cycle. J Gen Microbiol. 1988 May;134(5):1155–1163. doi: 10.1099/00221287-134-5-1155. [DOI] [PubMed] [Google Scholar]
- Sargent M. G., Bennett M. F. Amplification of a major membrane-bound DNA sequence of Bacillus subtilis. J Bacteriol. 1985 Feb;161(2):589–595. doi: 10.1128/jb.161.2.589-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent M. G. How do bacterial nuclei divide? Microbiol Sci. 1985 Aug;2(8):235–239. [PubMed] [Google Scholar]
- Saunders N. A., Harrison T. G., Kachwalla N., Taylor A. G. Identification of species of the genus Legionella using a cloned rRNA gene from Legionella pneumophila. J Gen Microbiol. 1988 Aug;134(8):2363–2374. doi: 10.1099/00221287-134-8-2363. [DOI] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986 May;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]