Abstract
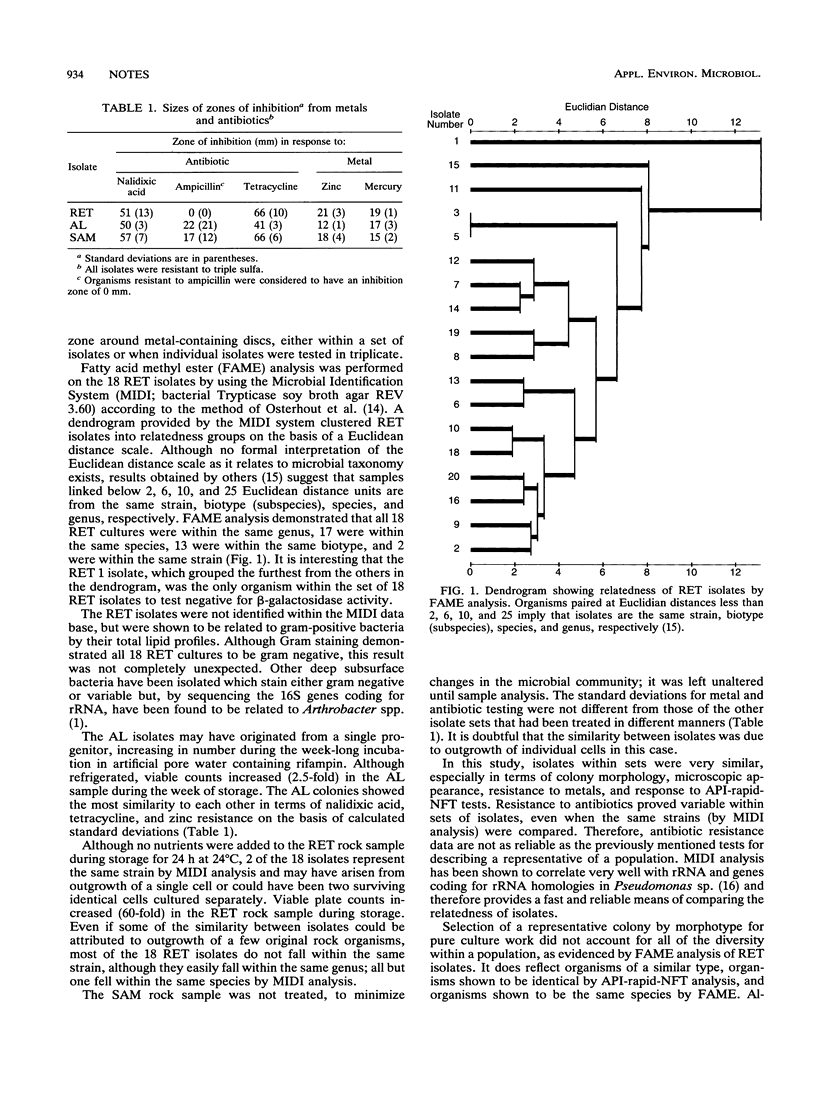
Sets of bacterial isolates with the same colony morphologies were selected from spread plates of bacteria from deep subsurface rock samples; each set had a unique morphology. API-rapid-NFT analysis revealed that isolates within a set were the same. Fatty acid methyl ester analysis of one set of isolates clustered organisms within the same species, defining variation between isolates at the biotype (subspecies) and strain levels. Metal resistances consistently tracked with colony morphology, while antibiotic resistances were less reliable.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amy P. S., Haldeman D. L., Ringelberg D., Hall D. H., Russell C. Comparison of Identification Systems for Classification of Bacteria Isolated from Water and Endolithic Habitats within the Deep Subsurface. Appl Environ Microbiol. 1992 Oct;58(10):3367–3373. doi: 10.1128/aem.58.10.3367-3373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill D. L., Fredrickson J. K., Thomas J. M. Vertical and horizontal variations in the physiological diversity of the aerobic chemoheterotrophic bacterial microflora in deep southeast coastal plain subsurface sediments. Appl Environ Microbiol. 1989 May;55(5):1058–1065. doi: 10.1128/aem.55.5.1058-1065.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Geesey G. G., Cheng K. J. How bacteria stick. Sci Am. 1978 Jan;238(1):86–95. doi: 10.1038/scientificamerican0178-86. [DOI] [PubMed] [Google Scholar]
- Fredrickson J. K., Balkwill D. L., Zachara J. M., Li S. M., Brockman F. J., Simmons M. A. Physiological diversity and distributions of heterotrophic bacteria in deep cretaceous sediments of the atlantic coastal plain. Appl Environ Microbiol. 1991 Feb;57(2):402–411. doi: 10.1128/aem.57.2.402-411.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson J. K., Hicks R. J., Li S. W., Brockman F. J. Plasmid incidence in bacteria from deep subsurface sediments. Appl Environ Microbiol. 1988 Dec;54(12):2916–2923. doi: 10.1128/aem.54.12.2916-2923.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez L. Molecular analysis of deep-subsurface bacteria. Appl Environ Microbiol. 1990 Jul;56(7):2108–2113. doi: 10.1128/aem.56.7.2108-2113.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhout G. J., Shull V. H., Dick J. D. Identification of clinical isolates of gram-negative nonfermentative bacteria by an automated cellular fatty acid identification system. J Clin Microbiol. 1991 Sep;29(9):1822–1830. doi: 10.1128/jcm.29.9.1822-1830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]


