Abstract
Alkylation of DNA at the O6-position of guanine is one of the most critical events leading to mutation, cancer, and cell death. The enzyme O6-methylguanine-DNA methyltransferase repairs O6-methylguanine as well as a minor methylated base, O4-methylthymine, in DNA. Mouse lines deficient in the methyltransferase (MGMT) gene are hypersensitive to both the killing and to the tumorigenic effects of alkylating agents. We now show that these dual effects of an alkylating agent can be dissociated by introduction of an additional defect in mismatch repair. Mice with mutations in both alleles of the MGMT gene and one of the mismatch repair genes, MLH1, are as resistant to methylnitrosourea (MNU) as are wild-type mice, in terms of survival, but do have numerous tumors after receiving MNU. In contrast to MGMT−/− MLH1+/+ mice with decrease in size of the thymus and hypocellular bone marrow after MNU administration, no conspicuous change was found in MGMT−/− MLH1−/− mice treated in the same manner. Thus, killing and tumorigenic effects of an alkylating agent can be dissociated by preventing mismatch repair pathways.
Chemical modification of cellular DNA can be induced by the action of alkylating substances, either externally administered or produced through endogenous metabolic pathways. Among the various alkylated bases formed in DNA, O6-methylguanine is regarded as being highly responsible for the induction of mutation and cancer in organisms (1–3). O6-methylguanine can pair with thymine as well as cytosine during DNA replication, leading to G:C to A:T transition mutation (4, 5).
To repair the premutagenic base in DNA, organisms possess a specific repair protein, O6-methylguanine-DNA methyltransferase. The enzyme transfers methyl groups from O6-methylguanine and O4-methylthymine, the latter being formed less frequently, in DNA to the cysteine residue of its own molecule, thereby repairing the DNA lesions in a single-step reaction. Because the reaction irreversibly inactivates the enzyme, the repair capacity for O6-methylguanine depends on the number of methyltransferase molecules in the cell (6–8). Some tumor-derived cell lines are hypersensitive to alkylating agents, and these cell lines, termed Mer− or Mex−, have little or no methyltransferase activity (9–11).
To clarify the roles of methyltransferase in carcinogenesis, animal models with altered levels of the enzyme activity have been developed. Transgenic mice carrying extra copies of methyltransferase genes and cDNA with functional promoters exhibited significantly reduced rates of tumor formation after administration of low doses of alkylating agents (12, 13). Recently, we established mouse lines defective in the MGMT gene, encoding O6-methylguanine-DNA methyltransferase (14). A large number of tumors occurred in MGMT−/− mice exposed to low doses of methylnitrosourea (MNU) and dimethylnitrosoamine whereas no or few tumors occurred in normal mice treated in the same manner (15, 16). Another notable feature of the gene-targeted mice was their extraordinary high sensitivity to killing effects of alkylating agents; LD50 of MGMT−/− mice was less than one-tenth of these values for both MGMT+/+ and MGMT+/− mice (14, 15). The bone marrow of the treated MGMT−/− mice became hypocellular, and there was a drastic decrease in the number of peripheral leukocytes and platelets, thereby indicating an impaired reproductive capacity of hematopoietic stem cells (14). Because methyltransferase apparently protected these mice from the pancytopenia caused by the alkylating agent, these alkylated bases may be responsible for death of the rapidly growing cells.
These lethal effects caused by simple alkylating agents may relate to inappropriate processing of mismatched bases. This was first suggested in studies with Escherichia coli dam− strains, in which mutations in the mismatch recognition genes mutS and mutL afforded protection against toxicity of alkylating agents (17, 18). Recent studies indicated that an acquired resistance of mammalian methyltransferase-deficient Mer− cell lines to alkylating agents is associated with the loss of capacity for mismatch repair, a phenomenon known as “methylation tolerance” (19, 20). The accumulation of alkylated bases in chromosomal DNA may provoke abortive mismatch repair, an event that could lead to cell death (21). If this thesis is tenable, then introduction of a mismatch repair gene defect would render methyltransferase-deficient mice resistant to the lethal action of alkylating agents. In such mice numerous tumors would occur on exposure to sublethal doses of alkylating agents because mutation-evoking methylated bases persist in the DNA.
To test this hypothesis, we generated mice defective in both MGMT and MLH1, the latter encoding a protein that functions at an early step of mismatch repair process. These MGMT−/− MLH1−/− mice exhibited an acquired resistance to MNU in terms of killing effect, but maintained the high sensitivity in tumor formation. Such mice may be useful for evaluating carcinogenic effects of various substances, including those for therapeutic application.
MATERIALS AND METHODS
Generation of MGMT−/− MLH1−/− Mice.
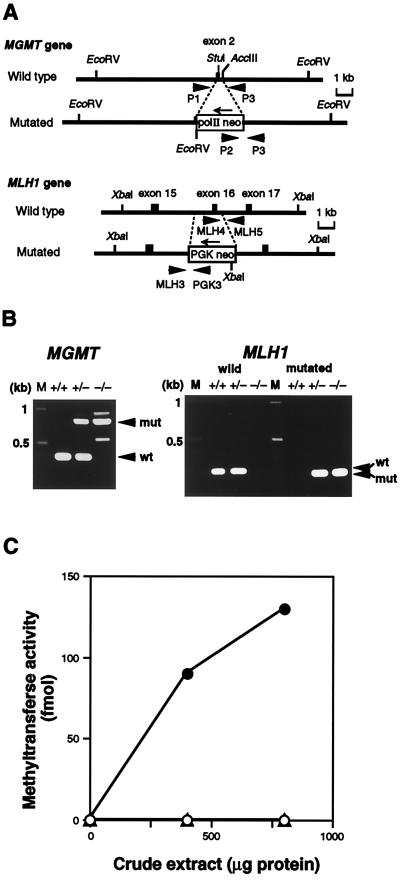
MGMT-knockout mice were developed as described (14). In brief, the targeting vector containing an 8.0-kb MGMT genomic sequence interrupted by a polII-neo-poly(A) cassette and flanked by a pair of herpes simplex thymidine kinase genes for negative selection (Fig. 1A Upper) was electroporated into CCE embryonic stem cells. Colonies doubly resistant to G418 and ganciclovir were selected and the ES cells were microinjected into C57BL/6J blastocysts to produce chimeric mice. Male chimeric mice were tested for germ-line transmission of the mutant allele and were then used to acquire heterozygous mutant mice. Homozygous mutant mice were obtained by crossing a pair of heterozygous mice. Development of MLH1+/– mutant mice used for these experiments will be described elsewhere (S.T., H.T., and T.N., unpublished data). A region of the MLH1 sequence carrying an exon corresponding to exon 16 of the human gene was replaced by a PGK-neo-poly(A) cassette (Fig. 1A Lower). This MLH1 mutation is an in-frame deletion of 165 nt, which is found in some Finnish HNPCC (hereditary nonpolyposis colorectal cancer) kindreds (22). The two types of gene-targeted mice were mated to produce MGMT−/− MLH1−/− mice. Genotypes of these mice were determined by PCR analyses and Southern blot hybridization, by using appropriate primers and probes (14). Primers for amplifying the MGMT sequence were P1 (5′-GTGTTGGACAGCCCTTTG-3′), P2 (5′-TGCAATCCATCTTGTTCAATG-3′), and P3 (5′-CTCATGGGATTCAACACC-3′), resulting in a 380-bp PCR product for wild-type allele and an 800-bp product for mutated allele. Primers for the wild-type MLH1 sequence were MLH4 (5′-AAGAAGAAAGCGGAGATGCTTGCAGAC-3′) and MLH5 (5′-GATAGATACATGCTGCTTCTGAGGGGA-3′), resulting in a 260-bp PCR product. For the mutated MLH1 allele, the primers used were PGK3 (5′-CCTGAAGAACGAGATCAGCAGCCTC-3′) and MLH3 (5′-GAACAGTCTGAGCGTGAAGGTTTCATG-3′), resulting in a 220-bp product (Fig. 1B).
Figure 1.
Targeted disruption of the mouse MGMT and MLH1 genes. (A) Schematic representation of targetings of the MGMT and MLH1 genes. Structures of parts of the wild-type (Upper) and the mutated sequences resulting from homologous recombination (Lower) are shown. Exons are shown as solid boxes. Thick arrows indicate PCR primers for genotyping of mice, and thin arrows represent the directions of transcription of the inserted cassettes. Appropriate restriction sites are shown. (B) Genotyping of progenies. To detect wild-type and mutated MGMT alleles (Left), PCR was done using three primers: P1, P2, and P3. The lengths of PCR products for wild-type (wt) and mutated (mut) alleles were 380 bp and 800 bp, respectively. For analyses of the MLH1 locus (Right), two sets of primers, MLH4-MLH5 (for wild type) and MLH3-PGK3 (for mutated allele), were used. The former gave a 260-bp PCR product (wt), and the latter a 220-bp one (mut). M, DNA size marker. (C) O6-methylguanine-DNA methyltransferase activity. Crude extracts were prepared from thymus, and the extracts were incubated with [3H]MNU-treated calf thymus DNA. (○), MGMT−/− MLH1+/+; (▵), MGMT−/− MLH1−/−; (•), MGMT+/+ MLH1+/+.
Assay of Methyltransferase Activity.
The activity was determined as described (23), but with slight modifications. Thymi of mice were broken into pieces in liquid nitrogen and suspended in buffer B (50 mM Tris⋅HCl, pH 7.5/10% glycerol/0.1 mM EDTA/1 mM DTT) containing 100 mM NaCl (24). The suspension was sonicated and centrifuged to collect the supernatant, as crude extract. The extract was incubated in 200 μl of 70 mM Hepes-KOH, pH 7.8/1 mM DTT/5 mM EDTA containing [3H]MNU-treated calf thymus DNA (2,750 Bq per assay) at 37°C for 15 min. [3H]MNU (17.5 Ci/mmol; 1 Ci = 37 GBq) was purchased from Amersham and used to prepare labeled alkylated DNA. After hydrolyzing the DNA in heated trichloroacetic acid, the methyl-accepted protein was collected by centrifugation and radioactivity was determined in a liquid scintillation counter.
MNU Administration.
To examine the susceptibility to an alkylating agent, 6-week-old mice were given an i.p. injection of MNU and survivors were counted at 30 days after the treatment. MNU (Nacalai Tesque, Kyoto, Japan) was dissolved in PBS immediately before use. Thymus and bone marrow were examined 7 days after treatment, and MNU-induced tumorigenesis was observed 8 weeks after administration.
RESULTS
Generation of MGMT−/− MLH1−/− Mice.
Using gene targeting techniques, we generated mice deficient in O6-methylguanine-DNA methyltransferase activity (14). MLH1 gene-knockout mice were developed by replacing the genomic DNA sequence containing an exon corresponding to exon 16 of the human MLH1 gene and the surrounding intron regions by a PGK/neo cassette (S.T., H.T., and T.N., unpublished data) (Fig. 1A). Doubly defective mice, MGMT−/− MLH1−/−, were obtained by crossing the two types of knockout mice. PCR analyses confirmed that these animals indeed carried replacement mutations in both alleles of the two genes (Fig. 1B). To ensure that the MGMT−/− MLH1−/− mice had no repair capacity for O6-methylguanine, the methyltransferase activity was determined. As shown in Fig. 1C, practically no methyltransferase activity was present in thymus samples of the double-mutant mice.
Susceptibility to MNU.
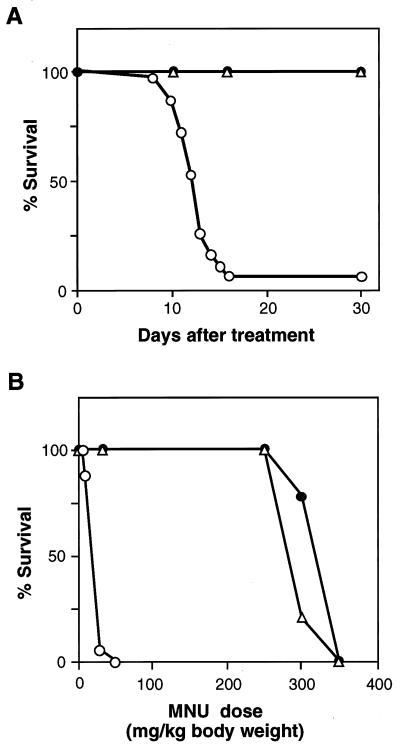
As an alkylating agent we have chosen MNU because it alkylates DNA without metabolic activation, yielding O6-methylguanine and O4-methylthymine at levels of 5–11% and 0.1–0.7% of the total alkylated DNA adducts, respectively, both in vitro and in vivo (25). Four groups of mice with different genotypes, each group consisting of about 40 animals (6 weeks old), were given a single i.p. injection of MNU (30 mg/kg of body weight). As a control, PBS was injected into these mice, all of which survived during the period of observation (over 30 days). As shown in Fig. 2A, 95% of MGMT−/− MLH1+/+ mice died within 17 days whereas MGMT+/+ MLH1+/+ and MGMT+/+ MLH1−/− mice, which carry normal levels of methyltransferase activity, survived for at least 30 days after treatment.
Figure 2.
Survival curves of mice given MNU treatment. (A) Duration of survival. Thirty-seven of MGMT−/− MLH1+/+, 37 of MGMT+/+ MLH1−/−, 38 of MGMT−/− MLH1−/−, and 36 of MGMT+/+ MLH1+/+ mice (each 6 weeks old) were given MNU (30 mg/kg of body weight) i.p. in the sixth postnatal week. These mice were observed for 30 days after the treatment. (B) Survival of mice given various doses of MNU. Six-week-old mice were given various doses of MNU i.p. For each dose 7 to 12 mice were used and data on survivors at 30 days after the administration were plotted. (○) MGMT−/− MLH1+/+; (▵), MGMT−/− MLH1−/−; (•), MGMT+/+ MLH1+/+ and MGMT+/+ MLH1−/− mice.
Of interest is the observation that all of MGMT−/− MLH1−/− mice survived after MNU treatment despite being completely devoid of methyltransferase activity. This was further demonstrated in an additional experiment, in which different doses of MNU were given to these mice (Fig. 2B). MGMT−/− MLH1−/− mice were as resistant to MNU as MGMT+/+ MLH1+/+ mice, in which the LD50 was about 280 mg/kg body weight. On the other hand, the mice defective in the methyltransferase gene alone (MGMT−/− MLH1+/+) were far more sensitive to MNU, and here the LD50 was about 20 mg/kg of body weight, that is, less than one-tenth of the value for the double-defective mice and the wild-type mice.
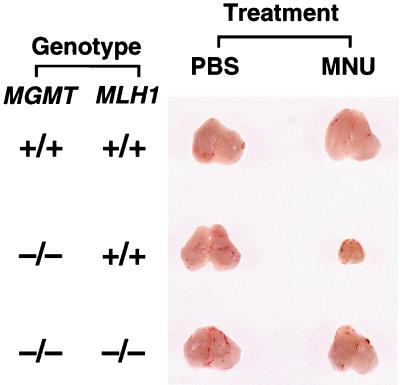
There was a remarkable reduction in body weight of the MNU-administered MGMT−/− MLH1+/+ mice (14). Reduction in size of certain organs, such as thymus and spleen, was evident. Fig. 3 shows the gross appearance of thymi of mice on the seventh day after exposure to MNU. However, these events in methyltransferase-deficient mice were nil in MGMT−/− MLH1−/− mice treated in exactly the same manner. Weight of the doubly defective mice was practically normal as was size of internal organs, even after treatment with MNU.
Figure 3.
Morphological comparison of the mouse thymus in the case of MNU treatment. Six-week-old mice with different genotypes were treated with MNU or PBS, and 8 weeks after this administration, the thymus of each mouse was excised. More than 10 mice were used for each group, and typical results are shown.
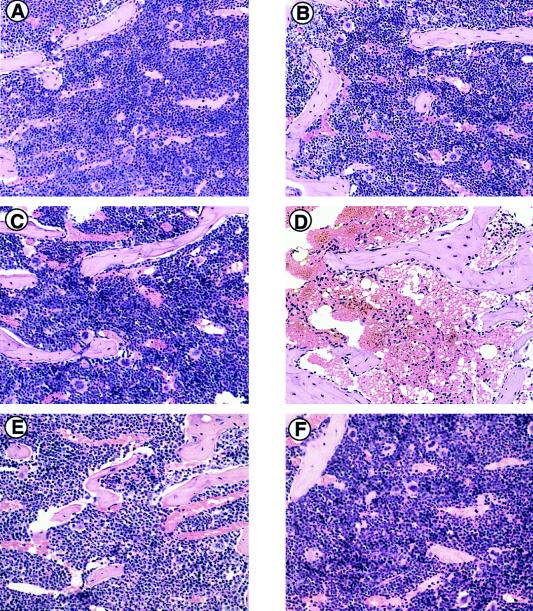
Fig. 4 shows the histology of findings in bone marrow from all the mice, given or not given MNU treatment. There is no apparent difference in the three groups of PBS-treated mice (Fig. 4 A, C, and E). Hematopoietic cells in the bone marrow of MGMT−/− MLH1+/+ mice treated with MNU were few (Fig. 4D), whereas various types of hematopoietic cells were preserved in similarly treated wild-type and MGMT−/− MLH1−/− mice (Fig. 4 B and F). Therefore, in this respect also, the doubly defective mice are resistant to MNU.
Figure 4.
Histopathological analyses of MNU-administered mice. (A–F) Microscopic observation of bone marrow of mice treated with PBS (A, C, and E) or MNU (B, D, and F). Six-week-old mice were given PBS or 30 mg/kg of MNU i.p. and were killed 7 days after treatment to search for changes in bone marrow. The paraffin-embedded sections were stained with hematoxylin and eosin. Original magnification in A–F was ×66. (A and B) MGMT+/+ MLH1+/+; (C and D) MGMT−/− MLH1+/+; (E and F) MGMT−/− MLH1−/− mice.
Tumor Formation After MNU Administration.
Methyltransferase-deficient mice would be less refractory to tumor-inducing effects of alkylating agents than methyltransferase-proficient mice, and this was clearly shown in our previous studies (15, 16). However, in these experiments, only small doses of alkylating carcinogens were given because methyltransferase-deficient mice are hypersensitive to the lethal effect of alkylating agents. The present finding that MGMT−/−MLH1−/− mice are as resistant as wild-type mice to MNU presents a unique opportunity to examine the suppressive effect of methyltransferase on tumor induction by alkylating carcinogens.
For this, MGMT+/+ MLH1+/+, MGMT+/+ MLH1−/−, and MGMT−/− MLH1−/− mice were given a single i.p. injection of MNU (30 mg/kg of body weight) 6 weeks postnatally, and organs of these animals were examined 8 weeks after MNU administration (Table 1). About 70% of the MGMT−/− MLH1−/− mice had a thymic lymphoma whereas no lymphoma was present in wild-type mice. The tumor covering lungs and heart with effusion into the thoracic cavity weighed 0.457 g on average and was about 10 times heavier than that of the normal thymus. In an about half the number of tumor-bearing animals, the lymphoma had infiltrated other organs. Histological examination revealed that the tumor was composed of a diffuse proliferation of lymphoma cells. In addition, lung adenomas were present in two of the MNU-administered MGMT−/− MLH1−/− mice.
Table 1.
Thymic lymphoma formed after exposure to a low dose of MNU
| Genotype | Sex | Number of treated mice | Number of mice with thymic lymphoma |
|---|---|---|---|
| MGMT+/+ MLH1+/+ | F | 19 | 0 (0%) |
| M | 17 | 0 (0%) | |
| MGMT−/− MLH1−/− | F | 17 | 13* (76%) |
| M | 21 | 14 (67%) | |
| MGMT+/+ MLH1−/− | F | 22 | 9 (41%) |
| M | 15 | 1 (7%) |
Mice (6 weeks old) were given 30 mg/kg of body weight of MNU i.p. and killed 8 weeks later.
Three mice died with thymic lymphoma 31, 46, and 51 days after MNU treatment.
When MGMT+/+ MLH1−/− mice were treated with MNU, a thymic lymphoma was also induced (see Table 1). The number was fewer as compared with the case of MGMT−/− MLH1−/− mice but the lymphoma infiltration was similar. This event may also be caused by inappropriate processing in mismatch repair. It should be noted that even without MNU administration small numbers of tumors were formed in MGMT+/+ MLH1−/− mice, in agreement with previous studies (26). This number was not increased significantly in the case of MGMT−/− MLH1−/− mice background (data not shown).
DISCUSSION
Making use of gene targeting, we developed mouse lines defective in both alleles of the MGMT gene (14). Such mice are extraordinarily sensitive to alkylating agents. Pancytopenia, atrophy of the thymus and the spleen, hypocellular bone marrow, and degenerative change in intestinal endothelial cells all occur. Because stem cells of bone marrow and epithelium rapidly divide and apoptotic cell death can occur after G2/M arrest in the second cycle of cell proliferation, rapid death of stem cells in such tissues might lead to dysfunction of vital organs. Induction of apoptotic cell death by alkylating agents occurred in mouse embryonic cell lines deficient in methyltransferase (27).
We then asked how the persistence of O6-methylguanine in DNA causes apoptotic cell death. The alkylated base is a structurally minor DNA lesion and does not seem to block DNA replication (2, 6–8). The DNA replication fork proceeds over the site of O6-methylguanine, inserting either cytosine or thymine opposite the methylated base. The O6-methylguanine-thymine pair would cause a G:C to A:T transition mutation after the second cycle of DNA replication or alternatively be recognized by mismatch repair proteins (21, 28). In the latter case, excision of part of the newly replicated strand with the mismatched base is followed by a resynthesis, again incorporating thymine opposite O6-methylguanine. These cycling events, which lead to formation of single-stranded regions at the site of O6-methylguanine residues, will produce double-strand gaps when the DNA is replicated, an event eventually resulting in cell death (21, 28). It is predicted that loss of mismatch repair mechanisms will prevent this process, thus conferring tolerance to simple alkylating agents. The present finding that MGMT−/− MLH1−/− mice are as resistant to MNU as the wild-type mice, in terms of survival, accords with this hypothesis.
A correlation between acquired resistance to alkylating agents and loss of mismatch repair capacity was noted in mammalian cell lines (19, 20). More recently, mouse ES cells carrying homozygous mutations in the MSH2 gene, encoding a mismatch recognition protein, were seen to have an increased resistance to alkylating agents in the presence of O6-benzylguanine, an inhibitor of O6-methylguanine-DNA methyltransferase (29). However, in a lower eukaryote, Saccharomyces cerevisiae, defects in any genes functioning at early steps of mismatch repair did not render cells more tolerant to killing effects of alkylating agents (30, 31). On the other hand, E. coli dam− strains with mutations in mismatch repair genes showed an increased resistance to alkylating agents (17, 18). Thus, responses to DNA alkylation-induced killing appear to differ depending on the type of cells and their genetic backgrounds.
Given these circumstances, it is remarkable that MGMT−/− MLH1−/− mice are far more resistant to MNU than are MGMT−/− MLH1+/+ mice, in terms of survival. The mouse is composed of many types of cells with varied reproduction capacities, but the susceptibility of mice to an alkylating agent may depend on susceptibility of the most sensitive cells, namely populations of the most actively growing cells. In our present study, death of the methyltransferase-deficient mice was closely related to bone marrow damage and dysplastic mucosa of intestines together with crypt abscess, and introduction of the MLH1 mutation resulted in disappearance of this myelosuppression.
In this way, the mismatch repair system appears to eliminate cells with potentially mutation-evoking DNA damage. This means that MGMT−/− MLH1−/− mice, if exposed to a low dose of alkylating agent, would have many tumors. This is indeed the case, as shown in the present study. Most of the doubly deficient mice had thymic lymphoma whereas no tumor was found in the wild-type mice, although the two types of mice showed similar degrees of resistance to an alkylating agent, in terms of survival. Thus, the killing and the tumorigenic effects of alkylating agents could be dissociated by disruption of the two genes related to independent DNA repair processes.
Although mutations in both alleles of the two genes might be a rare occurrence, expression of the genes could be suppressed under certain physiological conditions. In some tumor-derived cells, no or only little methyltransferase protein and its mRNA are formed (7, 8), and this lack of gene expression has been attributed to alterations in the cytosine methylation pattern of the promoter region of the gene (32–34). The lack of MLH1 expression was also seen to correlate with cytosine methylation of the promoter region (35). The absence of both MGMT and MLH1 expression might occur in certain cells within the human body, perhaps with important clinical implications. It has been well established that hereditary nonpolyposis colorectal cancer (HNPCC) is caused by a defect in mismatch repair genes, which is frequently associated with microsatellite instability. This type of defect can be seen in many types of sporadic tumors, not limited to colorectal cancer (36, 37). In such cases, application of carcinostatic drugs with an alkylation capacity would cause accumulation of mutations, which in turn convert the cell into a more malignant one. Thus, detailed characterization of tumor cells may be essential when prescribing chemotherapeutic agents.
Acknowledgments
We thank Y. Nakabeppu, T. Iwakuma, and H. Igarashi for discussion, M. Katsuki and K. Nakao for instruction in gene targeting experiments, and M. Ohara for helpful comments. This work was supported by grants from the Ministry of Education, Science, Sports and Culture, Japan, and from Uehara Memorial Foundation.
ABBREVIATION
- MNU
N-methyl-N-nitrosourea
References
- 1.Loveless A. Nature (London) 1969;223:206–207. doi: 10.1038/223206a0. [DOI] [PubMed] [Google Scholar]
- 2.Strauss B, Scudiero D, Henderson E. In: Molecular Mechanisms for Repair of DNA, Part A. Hanawalt P C, Setlow R B, editors. New York,: Plenum; 1975. pp. 13–24. [Google Scholar]
- 3.Sukumar S, Notario V, Martin-Zanca D, Barbacid M. Nature (London) 1983;306:658–661. doi: 10.1038/306658a0. [DOI] [PubMed] [Google Scholar]
- 4.Coulondre C, Miller J H. J Mol Biol. 1977;117:577–606. doi: 10.1016/0022-2836(77)90059-6. [DOI] [PubMed] [Google Scholar]
- 5.Loechler E L, Green C L, Essigmann J M. Proc Natl Acad Sci USA. 1984;81:6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- 7.Pegg A E. Cancer Res. 1990;50:6119–6129. [PubMed] [Google Scholar]
- 8.Sekiguchi M, Nakabeppu Y, Sakumi K, Tsuzuki T. J Cancer Res Clin Oncol. 1996;122:199–206. doi: 10.1007/BF01209646. [DOI] [PubMed] [Google Scholar]
- 9.Day R S, III, Ziolkowski C H J, Scudiero D A, Meyer S A, Mattern M R. Carcinogenesis. 1980;1:21–32. doi: 10.1093/carcin/1.1.21. [DOI] [PubMed] [Google Scholar]
- 10.Sklar R, Strauss B. Nature (London) 1981;289:417–420. doi: 10.1038/289417a0. [DOI] [PubMed] [Google Scholar]
- 11.Yarosh D B, Foote R S, Mitra S, Day R S., III Carcinogenesis. 1983;4:199–205. doi: 10.1093/carcin/4.2.199. [DOI] [PubMed] [Google Scholar]
- 12.Nakatsuru Y, Matsukuma S, Nemoto N, Sugano H, Sekiguchi M, Ishikawa T. Proc Natl Acad Sci USA. 1993;90:6468–6472. doi: 10.1073/pnas.90.14.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumenco L L, Allay E, Norton K, Gerson S L. Science. 1993;259:219–222. doi: 10.1126/science.8421782. [DOI] [PubMed] [Google Scholar]
- 14.Tsuzuki T, Sakumi K, Shiraishi A, Kawate H, Igarashi H, Iwakuma T, Tominaga Y, Zhang S, Shimizu S, Ishikawa T, et al. Carcinogenesis. 1996;17:1215–1220. doi: 10.1093/carcin/17.6.1215. [DOI] [PubMed] [Google Scholar]
- 15.Sakumi K, Shiraishi A, Shimizu S, Tsuzuki T, Ishikawa T, Sekiguchi M. Cancer Res. 1997;57:2415–2418. [PubMed] [Google Scholar]
- 16.Iwakuma T, Sakumi K, Nakatsuru Y, Kawate H, Igarashi H, Shiraishi A, Tsuzuki T, Ishikawa T, Sekiguchi M. Carcinogenesis. 1997;18:1631–1635. doi: 10.1093/carcin/18.8.1631. [DOI] [PubMed] [Google Scholar]
- 17.Jones M, Wagner R. Mol Gen Genet. 1981;184:562–563. doi: 10.1007/BF00352542. [DOI] [PubMed] [Google Scholar]
- 18.Karran P, Marinus M G. Nature (London) 1982;296:868–869. doi: 10.1038/296868a0. [DOI] [PubMed] [Google Scholar]
- 19.Branch P, Aquilina G, Bignami M, Karran P. Nature (London) 1993;362:652–654. doi: 10.1038/362652a0. [DOI] [PubMed] [Google Scholar]
- 20.Kat A, Thilly W G, Fang W-H, Longley M J, Li G-M, Modrich P. Proc Natl Acad Sci USA. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karran P, Bignami M. BioEssays. 1994;16:833–839. doi: 10.1002/bies.950161110. [DOI] [PubMed] [Google Scholar]
- 22.Papadopoulos N, Nicolaides N C, Wei Y-F, Ruben S M, Carter K C, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, et al. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 23.Nakabeppu Y, Kondo H, Kawabata S, Iwanaga S, Sekiguchi M. J Biol Chem. 1985;260:7281–7288. [PubMed] [Google Scholar]
- 24.Kawate H, Ihara K, Kohda K, Sakumi K, Sekiguchi M. Carcinogenesis. 1995;16:1595–1602. doi: 10.1093/carcin/16.7.1595. [DOI] [PubMed] [Google Scholar]
- 25.Beranek D T. Mutat Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 26.Baker S M, Plug A W, Prolla T A, Bronner C E, Harris A C, Yao X, Christie D-M, Monell C, Arnheim N, Bradley A, et al. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 27.Tominaga Y, Tsuzuki T, Shiraishi A, Kawate H, Sekiguchi M. Carcinogenesis. 1997;18:889–896. doi: 10.1093/carcin/18.5.889. [DOI] [PubMed] [Google Scholar]
- 28.Duckett D R, Drummond J T, Murchie A I H, Reardon J T, Sancar A, Lilley D M J, Modrich P. Proc Natl Acad Sci USA. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 30.Xiao W, Rathgeber L, Fontanie T, Bawa S. Carcinogenesis. 1995;16:1933–1939. doi: 10.1093/carcin/16.8.1933. [DOI] [PubMed] [Google Scholar]
- 31.Bawa S, Xiao W. Cancer Res. 1997;57:2715–2720. [PubMed] [Google Scholar]
- 32.Nakatsu Y, Hattori K, Hayakawa H, Shimizu K, Sekiguchi M. Mutat Res. 1993;293:119–132. doi: 10.1016/0921-8777(93)90063-m. [DOI] [PubMed] [Google Scholar]
- 33.Costello J F, Futscher B W, Tano K, Graunke D M, Pieper R O. J Biol Chem. 1994;269:17228–17237. [PubMed] [Google Scholar]
- 34.Qian X C, Brent T P. Cancer Res. 1997;57:3672–3677. [PubMed] [Google Scholar]
- 35.Kane M F, Loda M, Gaida G M, Lipman J, Mishra R, Goldman H, Jessup J M, Kolodner R. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 36.Kolodner R D. Trends Biochem Sci. 1995;20:397–401. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- 37.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]