Abstract
The modification of yeast artificial chromosomes through homologous recombination has become a useful genetic tool for studying gene function and enhancer/promoter activity. However, it is difficult to purify intact yeast artificial chromosome DNA at a concentration sufficient for many applications. Bacterial artificial chromosomes (BACs) are vectors that can accommodate large DNA fragments and can easily be purified as plasmid DNA. We report herein a simple procedure for modifying BACs through homologous recombination using a targeting construct containing properly situated Chi sites. To demonstrate a usage for this technique, we modified BAC clones containing the zebrafish GATA-2 genomic locus by replacing the first coding exon with the green fluorescent protein (GFP) reporter gene. Molecular analyses confirmed that the modification occurred without additional deletions or rearrangements of the BACs. Microinjection demonstrated that GATA-2 expression patterns can be recapitulated in living zebrafish embryos by using these GFP-modified GATA-2 BACs. Embryos microinjected with the modified BAC clones were less mosaic and had improved GFP expression in hematopoietic progenitor cells compared with smaller plasmid constructs. The precise modification of BACs through Chi-stimulated homologous recombination should be useful for studying gene function and regulation in cultured cells or organisms where gene transfer is applicable.
The introduction of transgenes into living embryos has had a profound affect on the study of gene function and regulation (1, 2). Traditionally, DNA constructs smaller than 20 kb are used for this purpose because of their accessibility to standard molecular manipulations. However, these smaller plasmid constructs are often missing important elements for the regulation of gene activity. With the development of artificial chromosomes capable of cloning DNA fragments up to 2 megabases long, it is now possible to use intact genomic loci as transgenes (3, 4).
Transgenic mice have been produced with yeast artificial chromosomes (YACs) containing a variety of gene loci (5, 6). Although wild-type YACs are often used for this purpose, the inherently high frequency of homologous recombination in yeast makes it possible to precisely modify YACs by deletion or insertion of exogenous DNA. This property can be exploited to precisely modify DNA sequences by using linear targeting constructs containing regions of homology with any YAC (7). These modifications can include insertion of a reporter gene or introduction of mutations that mimic genetic defects (8). Given the large cloning capacity of YACs (9), it is also possible to perform unique in vivo studies on gene regulation. However, despite the ease of modifying YACs, the production of transgenic animals with such large molecules of DNA is a formidable task. Methods aimed at improving both the yield and integrity of purified YAC DNA have been established (10, 11), yet it is still difficult to purify YAC DNA that is both structurally intact and at a concentration sufficient for many experimental applications including zebrafish transgenesis. Additionally, both modified and wild-type YACs can exist in a single yeast clone (4), which complicates purification and verification of the modified YAC.
Bacterial artificial chromosomes (BACs) are Escherichia coli F factor-based vectors capable of maintaining cloned DNA fragments up to 300 kb (12). Bacteriophage P1-derived artificial chromosomes (PACs) also are high-cloning-capacity vectors and are capable of maintaining inserts in the range of 100–300 kb (13). Because BACs and PACs are stably maintained in bacteria and are amenable to the same molecular manipulations as conventional plasmid DNA, they offer a significant advantage over working with YACs. Recently, the mouse circadian Clock gene was identified in transgenic mice by using a 140-kb BAC to rescue the Clock mutation (14). However, for BAC transgenic technology to be useful for studying gene function and regulation, methods for modifying these artificial chromosomes at precise locations must be established. A temperature-sensitive plasmid based system has been developed to produce gene replacements and deletions in the E. coli chromosome through homologous recombination (15) and was recently used to insert the lacZ marker gene into a BAC for the purpose of generating transgenic mice (16). This technique requires multiple recombination events to create the desired gene replacement and hybridization experiments must be performed to detect each one. A less cumbersome method of introducing DNA fragments into BACs through homologous recombination would be advantageous.
We describe herein an alternative method for modifying BACs in E. coli by using homologous recombination. To accomplish this, a linear DNA construct containing properly situated Chi sites [5′-GCTGGTGG-3′ (17)] was used to promote and target homologous recombination to BACs. In E. coli, properly oriented Chi sites located near each end of a linear DNA fragment stimulate transfer of the segment between them to homologous DNA by recombination through the RecBCD pathway (18). With this method, we modified BACs containing the zebrafish GATA-2 genomic locus by replacing the first coding exon with the green fluorescent protein (GFP) reporter gene. The Chi-stimulated recombination event occurred precisely without additional deletions or rearrangements of the BACs. We demonstrate that GATA-2 expression patterns in proliferating skin enveloping layer cells (EVL), the central nervous system, and hematopoietic progenitor cells can be recapitulated in living zebrafish embryos by using these GFP-modified GATA-2 BACs. This technology should facilitate the identification and examination of gene regulatory elements, not only in zebrafish but in any organism where gene transfer techniques are applicable.
METHODS
Creating a Targeting Construct for Homologous Recombination.
A plasmid, pRM4-N, containing triple Chi sites flanking a multiple cloning site (MCS) and NotI sites for linearization was constructed as follows. An oligonucleotide containing three adjacent Chi sites was used to introduce properly oriented Chi sites to the left of the MCS in plasmid pUC19 by using the Morph kit (5 Prime → 3 Prime); this destroyed the EcoRI site in the MCS and generated pRM1. Separately, triple Chi sites were introduced to the right of the MCS in pUC19, to generate pRM2; these MCS-proximal Chi sites are separated by the sequence 5′-CCA-3′, to generate an MscI restriction site used for identification of the insert. Appropriate restriction fragments from pRM1 and pRM2 were ligated to generate pRM3, with triple Chi sites on each side of the MCS, properly situated to stimulate recombination of any DNA fragment contained between them (17, 18). The origin of replication (ori)-containing BsaI–AflIII fragment of pRM3 was removed and substituted with the pBR322 ori-containing BsaI–PvuII fragment of pDA16 (18) to generate pRM4. The ori of pDA16 and pRM4 are flanked by EcoRI restriction sites. The ori-EcoRI fragment of pRM4 was removed by EcoRI digestion and replaced with a 700-bp ori fragment from pUC19 by using the PCR. Primers containing NotI restriction sites were used for this PCR to create an ori with a NotI site at each end. The vector and pUC19 ori DNA fragments were blunt-ended by using the Klenow fragment of E. coli DNA polymerase I and the ori fragment was treated with T4 polynucleotide kinase before ligation with T4 DNA ligase. Deletion of the EcoRI sites and addition of NotI sites were verified by restriction digestion.
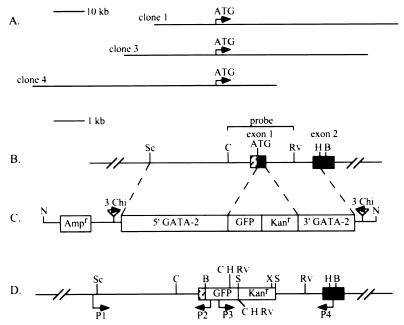
Zebrafish GATA-2 genomic fragments (19) containing 3.6 kb of DNA 5′ to the ATG initiation codon and 1.7 kb beginning at the first intron after the ATG of GATA-2 and extending into the second coding exon were ligated into the MCS of pRM4-N. The 1-kb GFP reporter gene (20) and 1.24-kb kanamycin-antibiotic-resistance gene from pUC4Km (21) were inserted between the two GATA-2 genomic fragments with the GFP gene adjacent to the 5′ GATA-2 fragment. This constitutes the 10.6-kb GATA-2 homologous recombination targeting construct (G2TC) used in this study (Fig. 1C). Recombination between a linearized G2TC DNA fragment and the GATA-2 gene will result in replacement of the 226-bp first coding exon with 2.24 kb representing the GFP and kanamycin genes.
Figure 1.
Schematic representation of the zebrafish GATA-2 genomic locus before and after modification by the Chi-containing G2TC DNA fragment. (A) Genomic structure of the three GATA-2 BAC clones and their relationships to the ATG initiation codon. Arrows indicate the orientation of the GATA-2 gene. (B) Region of the wild-type GATA-2 gene locus targeted for homologous recombination. (C) The nonreplicative G2TC DNA fragment containing Chi sites properly situated to promote homologous recombination (solid arrows). The edges of the 5′ and 3′ regions of homology with the GATA-2 gene locus are indicated by dashed lines. (D) GATA-2 gene locus after replacement of the first coding exon with the GFP and kanamycin genes. This modification adds several restriction sites to the GATA-2 gene as follows: B, BamHI; C, ClaI; Rv, EcoRV; H, HindIII; N, NotI; Sc, SacI; S, SalI; X, XhoI. Primers P1/P2 and P3/P4 were used in a PCR to identify BAC DNA with the correct gene replacement. Primers P1 and P4 were used to sequence the edges of the 5′ and 3′ regions of homology, respectively.
Preparation of GATA-2 BACs for Homologous Recombination.
A zebrafish genomic BAC library was screened with a 32P-labeled zebrafish GATA-2 cDNA probe (22) by following the protocol outlined by Genome Systems (St. Louis). In conjunction with restriction enzyme mapping of the BAC clones, we performed PCR using an Expand Long Template PCR system (Boehringer Mannheim) for 30 cycles (94°C for 10 s; 65°C for 30 s; 68°C for 10 min) to determine the extent of the 5′ and 3′ sequences. For the 5′ long PCR, we used three primers that bind 10 kb upstream of the GATA-2 ATG initiation codon. The 3′ long PCR was performed by using three primers that bind near the GATA-2 polyadenyltion site. These six primers were used in combination with either T7 or SP6 primers whose sequences are present in the BAC vector. BAC DNA was purified from DH10B bacteria by using Qiagen’s Plasmid Maxi kit and electrocompetent MC1061 bacteria (23) [F− araD139 Δ(ara-leu)7697 galU galK16 Δ(lac)X74 rpsL (Strr) hsdR2 (rk− mk+) mcrA mcrB1] were electroporated with the BAC DNA. Transformants were plated on Luria–Bertani (LB) agar plates containing chloramphenicol (12.5 μg/ml) and grown overnight at 37°C. The presence of intact BAC DNA in the MC1061 bacteria was verified by comparison of BAC DNA from chloramphenicol-resistant colonies with BAC DNA from the original DH10B bacterial strain after digestion with various restriction enzymes. For transformation with linearized G2TC DNA, MC1061 bacteria containing GATA-2 BAC DNA were made competent by using a CaCl2 method (24).
Transformation and Selection of Recombinants.
Linearized G2TC DNA was prepared by digesting G2TC plasmid with NotI overnight at 37°C and purifying the 9.9-kb targeting construct from the 700-bp ori by using a GENECLEAN kit (Bio 101). For each transformation, 200–400 ng of linearized G2TC DNA were diluted in 10 μl of MCT buffer (18), and 120 μl of CaCl2 competent MC1061 cells were added. The mixture was incubated on ice for 50 min, heat-shocked at 42°C for 1.5 min, and returned to ice for 2 min, and after the addition of 1 ml of LB medium, the bacteria were incubated at 37°C for 1 h with shaking. Cells were centrifuged at 4,000 × g for 2 min and 900 μl of LB was removed. The bacteria were resuspended in the remaining LB and plated on one LB agar plate containing kanamycin (50 μg/ml) and chloramphenicol (12.5 μg/ml). After overnight incubation at 37°C, selected colonies were grown overnight at 37°C in 10 ml of liquid LB containing kanamycin and chloramphenicol as described above. Aliquots from colonies that grew in liquid culture were streaked onto LB agar plates containing ampicillin (100 μg/ml) and incubated overnight at 37°C. Because the ampicillin-resistance gene is located outside of the regions of GATA-2 homology in the targeting construct (Fig. 1C), ampicillin resistance will be lost when homologous recombination occurs (Fig. 1D). Ampicillin-resistant colonies may arise from transformation with plasmid DNA or by nonhomologous recombination (18). Consequently, only ampicillin-sensitive colonies were further analyzed at the molecular level.
Molecular Analysis to Verify Homologous Recombination.
BAC plasmid DNA from kanamycin/chloramphenicol-resistant and ampicillin-sensitive colonies was prepared from 10-ml cultures by using a Wizard Plus Minipreps kit (Promega) by following the manufacturer’s recommendations for endA-positive bacterial strains. PCR was performed by using primers (Fig. 1D) designed to generate products spanning the region of GATA-2 homology. Primer P1 (5′-GGAGAATGATAAATGCGCGGTG-3′) binds 95 bp upstream of the 5′ GATA-2 fragment in the G2TC and primer P2 (5′-TTCCGTATGTTGCATCACCTTCACC-3′) binds 100 bp downstream from the GFP ATG initiation codon, creating a 5′ PCR product of 3.8 kb. Primer P3 (5′-AATGTATCAATCATGGCAGAC-3′) binds 470 bp downstream from the GFP ATG initiation codon and primer P4 (5′-CTGAGGAGGGTACCGGGATGAA-3′) binds the second coding exon of GATA-2 120 bp downstream of the 3′ GATA-2 fragment in the G2TC, creating a 3′ PCR product of 3.6 kb.
The modified BAC plasmid DNA was digested at unique restriction sites present within the GFP and kanamycin genes, and the resulting patterns were compared with those from wild-type BAC DNA by Southern blot analysis. A 2-kb wild-type ClaI–EcoRV GATA-2 genomic fragment (Fig. 1B), extending from the 5′ untranslated region to the first intron after the ATG, was used as a probe after labeling with [α-32P]dCTP by using a Random Primed DNA labeling kit (Boehringer Mannheim). Parts of the 5′ and 3′ regions of homology between the G2TC and the wild-type BACs were sequenced by using primers P1 and P4 described above. One modified BAC clone representing each of the three GATA-2 BAC clones was sequenced.
Microinjection and Analysis of Zebrafish Embryos.
Modified BAC plasmid DNA was prepared from 500-ml LB cultures containing kanamycin and chloramphenicol as described above with Qiagen’s Plasmid Maxi kit. Two 70% ethanol washes were performed and the DNA was resuspended in 100 μl of 1× TE (10 mM Tris/1 mM EDTA, pH 8.0). For microinjection of BAC plasmid, the DNA was diluted to 80–120 μg/ml in sterile double distilled H2O, making the final TE concentration approximately 0.5×, and KCl was added to a final concentration of 100 mM. Linearized BAC DNA was prepared for microinjection by digesting BAC plasmid with NotI to release the genomic insert from the BAC vector followed by phenol/chloroform extraction, 1:1 (vol/vol), precipitation, and dilution as described above. Single-cell-stage wild-type zebrafish embryos were microinjected with either plasmid or linearized BAC DNA and maintained as described (25). Microinjected embryos were examined under a fluorescein isothiocyanate filter (filter set 09; excitation BP 450–490 nm; beamsplitter FT 510; emission LP 520 nm) on a Zeiss microscope equipped with a video camera system as described (19). Each modified BAC was injected independently 2–5 times and the data obtained were pooled. The expression pattern of GATA-2 was determined by whole-mount RNA in situ hybridization as described (19).
RESULTS
Genomic Structure of Zebrafish GATA-2 BAC Clones.
We have chosen the GATA-2 gene to test the feasibility of modifying BACs through homologous recombination because it has a complex, but very specific, expression pattern during early zebrafish development. A BAC zebrafish genomic library was screened for the GATA-2 gene and four positive clones were identified. Pulsed-field gel electrophoresis of NotI-digested BAC DNA revealed that all four clones contained inserts of 70–80 kb. Our restriction enzyme analysis suggested that two clones were similar in their genomic structures. Therefore, only three BAC clones were used for further studies. With 5′ or 3′ long PCR, we determined that GATA-2 BAC clone 1 contained approximately 20 kb of 5′ sequence and that BAC clone 4 had greater than 55 kb of 5′ sequence. Restriction enzyme mapping suggested that BAC clone 3 contained approximately 30 kb of 5′ sequence. The three overlapping BAC clones and their relationships to the GATA-2 ATG initiation codon are illustrated in Fig. 1A.
Homologous Recombination with BACs.
The BAC plasmids were originally propagated in E. coli strain DH10B. Because this strain is mutated in a gene (rec−) required for homologous recombination (26), we transformed recombination proficient bacteria with the GATA-2 BAC clones. Strain MC1061, a rec+ strain, was chosen for the initial transformations. After electroporation, we performed restriction digestions of BAC plasmid DNA from independent clones to assure that no detectable structural changes had occurred in the MC1061 cells. Only those bacteria containing BACs that appeared to be identical to the original BAC clones from strain DH10B were used for gene targeting experiments. Modified BACs were maintained in strain MC1061 and we found no evidence to indicate that rearrangements of the BAC DNA had occurred.
We have used a linear targeting construct (Fig. 1C) containing three Chi sites on each side of the DNA that shares homology with the GATA-2 BAC clones. When strain MC1061 bacteria containing GATA-2 BACs were transformed with linearized G2TC DNA, a variable number of kanamycin/chloramphenicol-resistant colonies were obtained. On average, we obtained 20 colonies per transformation with 200–400 ng of linearized G2TC DNA. Although 14 of 84 (17%) of the kanamycin/chloramphenicol-resistant transformant colonies analyzed by ampicillin selection were ampicillin-sensitive, 11 of 14 (79%) of these colonies had the correct gene replacement (Table 1). Because eight independent transformation experiments were performed, this represents a frequency of 2.8 gene replacements per transformation with 200–400 ng of G2TC DNA. The structure of the GATA-2 genomic locus before and after homologous recombination with linearized G2TC DNA is shown in Fig. 1 B and D, respectively.
Table 1.
Chi-stimulated gene targeting to GATA-2 BACs in recombination-proficient bacteria
| BAC clone | Transformation experiments, n | Colonies analyzed by ampicillin selection, n | Ampicillin, sensitive colonies, n | Gene targeting events, n |
|---|---|---|---|---|
| 1 | 4 | 58 | 8 | 7 |
| 3 | 2 | 14 | 4 | 2 |
| 4 | 2 | 12 | 2 | 2 |
| Total | 8 | 84 | 14 | 11 |
Verifying Homologous Recombination at the Molecular Level.
PCR was used to initially identify recombined BAC plasmid DNA. The predicted 3.8-kb and 3.6-kb fragments spanning the 5′ and 3′ regions of G2TC homology, respectively, were obtained from 11 of 14 of the ampicillin-sensitive colonies (data not shown). A comparison of restriction digestion patterns from these 11 modified BACs revealed that they were indeed different from their wild-type counterparts. Furthermore, all of the modified clones, representing each of the GATA-2 BACs, had the expected restriction patterns, suggesting that they were modified identically.
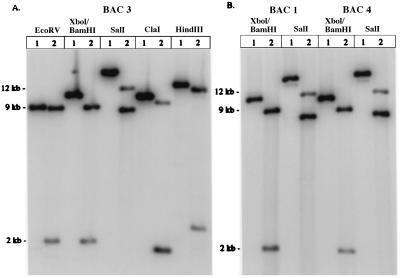
Southern blot analysis was performed on one modified clone from each of the original three GATA-2 BACs. We used the wild-type ClaI–EcoRV GATA-2 genomic fragment (Fig. 1B) as a probe because it contains sequences lying on either side of the inserted GFP and kanamycin genes. If homologous recombination occurs and the GFP and kanamycin genes replace the first coding exon, then several new restriction sites will be added to this region of the gene (Fig. 1D). These sites are unique to the modified BACs and provide a means of comparing them with the corresponding wild-type BACs. Restriction digestion of BAC clone 3 with six different enzymes demonstrated that the modification occurred as predicted (Fig. 2A). BAC clones 1 and 4 were also shown, by both XhoI/BamHI and SalI restriction enzyme digestion (Fig. 2B), to be modified correctly. In each case, a single large wild-type restriction fragment was cleaved into smaller fragments because of the inserted GFP and kanamycin genes. The size predictions for these smaller fragments were based on previous mapping and sequence analyses of the GATA-2 genomic locus (19) and the GFP and kanamycin (21) genes. As a further test that the gene replacement was fully homologous, without the formation of additional deletions or rearrangements, we sequenced 800 bp spanning the edges of homology between the G2TC DNA and the GATA-2 gene locus by using primers P1 and P4 (Fig. 1D). When compared with the wild-type GATA-2 genomic locus, no deletions or rearrangements were found at these locations (data not shown).
Figure 2.
Southern blot analysis of three different GATA-2 BAC clones modified by the G2TC DNA fragment. Newly introduced restriction sites (Fig. 1D) were used to digest both wild-type (lanes 1) and modified (lanes 2) BAC DNA. (A) GATA-2 BAC clone 3 was digested with the indicated enzymes and probed with a 2-kb wild-type ClaI–EcoRV genomic fragment (Fig. 1B). A 9-kb wild-type EcoRV fragment became 8.8-kb and 2-kb fragments. An 11-kb wild-type XhoI–BamHI fragment became 9-kb and 2-kb fragments. A 20-kb wild-type SalI fragment became 12-kb and 8.6-kb fragments. A 10.5-kb wild-type ClaI fragment became 9.5-kb and 1.7-kb fragments. A 13.5-kb wild-type HindIII fragment became 12-kb and 2.5-kb fragments. GATA-2 BAC clones 1 and 4 (B) also contained the desired gene replacement.
GFP Expression in Living Zebrafish Embryos Microinjected with Modified GATA-2 BAC DNA.
Through microinjection of smaller plasmid DNA constructs, we have previously demonstrated that GFP can be used to monitor GATA-2 enhancer/promoter activities in multiple tissues of living zebrafish embryos (19). This provides a reference to determine whether a GFP-modified GATA-2 BAC can recapitulate the expression pattern of GATA-2 during early development of zebrafish. We chose to purify BAC plasmid DNA for microinjection by using a Qiagen kit because reported methods for purifying artificial chromosomes (10, 16) produce DNA at a concentration too low for zebrafish transgenesis. With this method, we were able to obtain greater than 20 μg of plasmid DNA from a 500-ml liquid culture. To assure that BAC DNA remained intact throughout the microinjection procedures, DNA was first loaded into and ejected from a microinjection needle and analyzed by pulsed-field gel electrophoresis. Minimal shearing of either plasmid or NotI-linearized BAC DNA occurred after this procedure (data not shown).
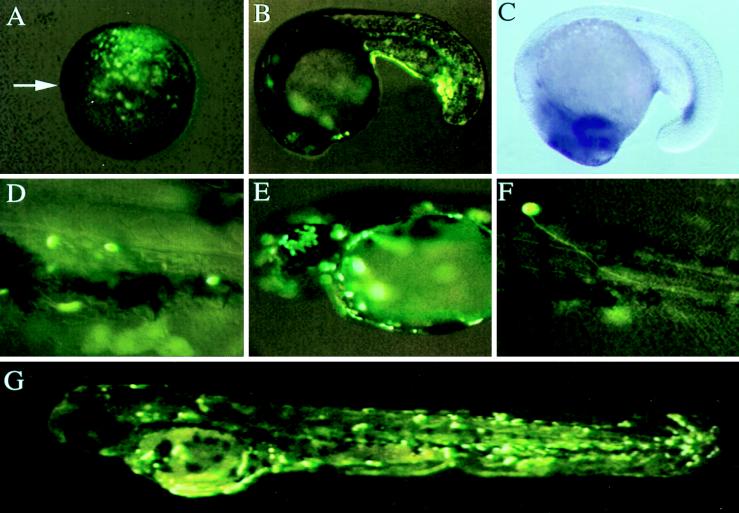
GFP reporter gene expression in microinjected embryos was examined at a number of distinct developmental stages by using fluorescence microscopy. For all three BAC clones, GFP expression was observed in the superficial EVL cells beginning approximately 4 h after injection (data not shown). By the shield stage (6 h), GFP-positive cells were observed in the ventral ectoderm and mesoderm (Fig. 3A). Analogous with our previous results with 7.3-kb (P1-GM2) and 4.7-kb (P2-GM2) GATA-2 promoter/GFP constructs (19), modified GATA-2-BAC-injected embryos developed GFP-positive hematopoietic cells (Fig. 3 B and D), proliferating skin EVL cells (Fig. 3 E and G), and neurons (Fig. 3F). These expression patterns were identical when either plasmid or NotI-linearized BAC DNA was used. In contrast, when linearized G2TC DNA was microinjected, only proliferating skin EVL cells and some hematopoietic cells displayed GFP fluorescence (data not shown). This indicated that additional enhancers required for neuronal expression of GATA-2 were provided by the BAC clones. Indeed, we know from our previous sequence analysis (19) that the G2TC DNA is missing a neuronal enhancer located at −3,812 bp from the GATA-2 ATG initiation codon. Because all of the injected GATA-2 BACs produced GFP expression in the central nervous system, we further conclude that the targeting construct recombined correctly with the GATA-2 BACs to produce the desired gene replacement.
Figure 3.
GFP expression from modified GATA-2 BACs in living zebrafish embryos. (A) GFP expression driven by modified BAC clone 4 in the ventral ectoderm and mesoderm of a 6-h shield stage embryo. Arrow indicates the dorsal shield. (B) GFP expression driven by modified BAC clone 4 in the posterior intermediate cell mass of a 20-h embryo. (C) GATA-2 expression pattern in an 18-h embryo as detected by RNA in situ hybridization. (D) GFP expression driven by modified BAC clone 3 in circulating hematopoietic cells of a 48-h embryo. GFP expression driven by modified BAC clone 4 in the EVL (E and G) and in a motoneuron (F) of 48-h embryos.
Compared with embryos injected with smaller DNA constructs that also produced GFP expression in skin EVL cells, neurons, and blood, embryos injected with BACs were less mosaic. GFP expression in the EVL cells was evenly distributed throughout the bodies of injected embryos (Fig. 3G). In some embryos injected with the modified BACs, GFP expression in the intermediate cell mass (Fig. 3B) was similar to the pattern of GATA-2 expression detected by RNA in situ hybridization (Fig. 3C) and was more extensive in the posterior intermediate cell mass than what we have observed with a 7.3-kb (P1-GM2) GATA-2 promoter construct (19). To assess GFP expression in hematopoietic cells, we examined circulating blood cells in 48-h embryos. Similar GFP expression patterns in hematopoietic cells were obtained with each of the three modified GATA-2 BAC clones (data not shown). This was indicated by the percentage of 48-h embryos possessing fluorescent circulating blood cells. We observed that 45% (n > 200) of modified BAC clone 4-injected embryos had GFP expression in circulating blood cells. This contrasts with the patterns observed with smaller constructs containing 7.3 kb or less of GATA-2 promoter where 1–15% (n > 1,000) of the embryos had GFP expression in circulating blood cells (19).
DISCUSSION
The ability to target homologous recombination to precise chromosomal locations has provided opportunities for the study of gene expression in whole animal systems (27). Modification of YACs through gene targeting and the generation of transgenic mice with these artificial chromosomes makes it possible to study many aspects of gene regulation during both normal development and the course of genetic disease (4). However, given the difficulties encountered when working with YAC DNA, we chose to develop a method whereby BACs can be modified through targeted homologous recombination for the purpose of gene expression studies in zebrafish. These artificial chromosomes can easily be purified intact and in a concentration suitable for analysis in zebrafish.
In this study we used a linear construct containing Chi sites to target homologous recombination in E. coli to BACs containing the zebrafish GATA-2 genomic locus. The result was precise replacement of the first coding exon of GATA-2 with the GFP and kanamycin genes. For the Chi-stimulated recombinational event to occur, a recombination-proficient bacterial strain possessing an intact RecBCD pathway was used. In this environment, a Chi-containing linear DNA molecule interacts with the RecBCD enzyme resulting in homologous recombination with the bacterial (18, 28, 29) or artificial chromosome (Table 1). The precision of the gene targeting event is an important aspect of this technique because it means that desired mutations and modifications can be introduced into artificial chromosomes with accuracy.
The chance that homologous recombination will occur at a particular chromosomal location is increased by Chi sites; however, this is still a rare event. Therefore, we used a positive–negative antibiotic selection procedure to efficiently select for bacterial colonies containing modified BACs. Positive selection identified those BAC-containing bacteria that integrated the kanamycin gene and negative selection identified bacteria that were ampicillin-sensitive. Through this enrichment process, we were able to minimize the number of bacterial colonies requiring molecular analyses, thereby saving both time and money. Molecular analyses of the 14 ampicillin-sensitive colonies revealed that 11 had undergone the correct gene targeting event. This represents a frequency of 2.8 gene replacements per transformation experiment using 200–400 ng of G2TC DNA. With other rec+ bacterial strains, Dabert and Smith (18) obtained average gene replacement frequencies of 1.4–6.4 per transformation with 100 ng of DNA when using a linear Chi-containing DNA construct to target the E. coli chromosome. Consequently, our results demonstrate that gene targeting to BACs using Chi-stimulated homologous recombination can occur with an efficiency similar to targeting the E. coli chromosome.
We used 3.6 kb and 1.7 kb of 5′ and 3′ homologous DNA sequence (Fig. 1C), respectively, to target recombination with the zebrafish GATA-2 genomic locus. Chi can function over several kilobases of heterologous sequence (30), and the minimum amount of homologous DNA required for this event to occur may be as little as 25 bp (31). The efficiency for this gene targeting event may be increased if larger regions of homology are used in the targeting construct. We have used this technique to modify the zebrafish rag1 gene contained within a BAC (data not shown). In this case, 4.7 kb of 5′ and 5 kb of 3′ homologous DNA sequences were used in the targeting construct. After one transformation experiment with the linearized targeting construct, we found that 14 of 15 (93%) of the kanamycin/chloramphenicol-resistant colonies were ampicillin-sensitive, and all 14 of these colonies had the desired gene replacement.
We chose zebrafish to demonstrate the usefulness of this technique for two reasons. (i) Our previous studies indicate that zebrafish provide an excellent system for analyzing gene enhancer/promoter activities using GFP because the developing embryo is transparent (19, 32). (ii) Artificial chromosomes have not been used to analyze gene expression in zebrafish. Previously, the GFP reporter gene has been used as part of DNA constructs less than 20 kb to study promoter elements in zebrafish (19, 32–35). Our results demonstrate that BACs can correctly drive the expression of a reporter gene in living zebrafish embryos. Specifically, embryos injected with GFP-modified GATA-2 BAC clones developed GFP expression in developing skin EVL cells, neurons, and circulating blood cells. We have reported that smaller plasmid constructs produced EVL expression when microinjected into zebrafish embryos (19). Because GATA-2 expression in EVL cells has not been reported in the literature, we hypothesized that a silencer element that would normally suppress EVL expression was missing from these smaller constructs. Given the extensive 5′ and 3′ sequences in the BAC clones, our results further suggest that GATA-2 is expressed in EVL cells. Furthermore, the percentage of BAC-injected 48-h embryos with GFP expression in hematopoietic cells was higher than previously reported for smaller GATA-2 promoter constructs (19). These results suggest that the GFP-modified GATA-2 BACs more accurately duplicated the GATA-2 expression pattern that occurs in hematopoietic cells. This difference was probably caused by the increased susceptibility of smaller plasmid constructs to position effects (36) but may be attributable to an enhancer element that is present in the BAC clones but absent from smaller constructs.
In summary, we have developed a simple procedure for modifying BACs in E. coli using Chi sites to promote homologous recombination between a linear DNA fragment and the artificial chromosomes. This method should also be adaptable to the modification of PACs. The ability to expediently modify these artificial chromosomes for transgenic experiments should facilitate in vivo genetic studies, not only in zebrafish but also in any organism amenable to transgenic methods. In addition to studying gene regulatory elements, modified BACs carrying mutations that mimic genetic lesions can be used to study genetic defects in whole animal systems. Recent chemical mutagenesis screens in zebrafish have generated more than 1,000 different mutants with defects in most developmental processes (37, 38). Our results indicate that DNA constructs microinjected into zebrafish embryos as part of large genomic fragments can display the correct spatiotemporal gene expression pattern. Given the availability of zebrafish BAC and PAC libraries, it will be possible to supplement positional cloning efforts at identifying genes responsible for these mutations by microinjecting artificial chromosomes into zebrafish embryos.
Acknowledgments
We thank Hong Tang, Billie Moore, Bruce Ong, and Carolyn Leithner for technical assistance and Rhea-Beth Markowitz for helpful discussions. This work was also supported in part by grants from the American Heart Association of Georgia (to S.L.) and from the National Institutes of Health (to G.R.S.). B.H.P. is the recipient of a Howard Hughes Medical Institute Postdoctoral Fellow for Physicians. L.I.Z. is an Associate Investigator of the Howard Hughes Medical Institute. S.L. is a recipient of The American Society of Hematology Scholar Award.
ABBREVIATIONS
- YAC
yeast artificial chromosome
- BAC
bacterial artificial chromosome
- GFP
green fluorescent protein
- PAC
bacteriophage P1-derived artificial chromosome
- EVL
enveloping layer
- MCS
multiple cloning site
- LB
Luria–Bertani
References
- 1.Hanahan D. Science. 1989;246:1265–1275. doi: 10.1126/science.2686032. [DOI] [PubMed] [Google Scholar]
- 2.Jaenisch R. Science. 1988;240:1468–1474. doi: 10.1126/science.3287623. [DOI] [PubMed] [Google Scholar]
- 3.Huxley C. Trends Genet. 1997;13:345–347. doi: 10.1016/s0168-9525(97)01256-0. [DOI] [PubMed] [Google Scholar]
- 4.Peterson K R, Peterson K R, Clegg C H, Li Q, Stamatoyannopoulos G. Genet Eng. 1997;19:235–255. [Google Scholar]
- 5.Forget B G. Proc Natl Acad Sci USA. 1993;90:7909–7911. doi: 10.1073/pnas.90.17.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schedl A, Ross A, Lee M, Engelkamp D, Rashbass P, van Heyningen V, Hastie N D. Cell. 1996;86:71–82. doi: 10.1016/s0092-8674(00)80078-1. [DOI] [PubMed] [Google Scholar]
- 7.Reeves R H, Pavan W J, Hieter P. Genet Anal Tech Appl. 1990;7:107–113. doi: 10.1016/0735-0651(90)90015-8. [DOI] [PubMed] [Google Scholar]
- 8.Peterson K R, Li Q L, Clegg C H, Furukawa T, Navas P A, Norton E J, Kimbrough T G, Stamatoyannopoulos G. Proc Natl Acad Sci USA. 1995;92:5655–5659. doi: 10.1073/pnas.92.12.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burke D T, Carle G F, Olson M V. Science. 1987;236:806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- 10.Schedl A, Larin Z, Montoliu L, Thies E, Kelsey G, Lehrach H, Schutz G. Nucleic Acids Res. 1993;21:4783–4787. doi: 10.1093/nar/21.20.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gnirke A, Huxley C, Peterson K, Olson M V. Genomics. 1993;15:659–667. doi: 10.1006/geno.1993.1121. [DOI] [PubMed] [Google Scholar]
- 12.Shizuya H, Birren B, Kim U J, Mancino V, Slepak T, Tachiiri Y, Simon M. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ioannou P A, Amemiya C T, Garnes J, Kroisel P M, Shizuya H, Chen C, Batzer M A, de Jong P J. Nat Genet. 1994;6:84–89. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- 14.Antoch M P, Song E J, Chang A M, Vitaterna M H, Zhao Y, Wilsbacher L D, Sangoram A M, King D P, Pinto L H, Takahashi J S. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X W, Model P, Heintz N. Nat Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- 17.Smith G R, Kunes S M, Schultz D W, Taylor A, Triman K L. Cell. 1981;24:429–436. doi: 10.1016/0092-8674(81)90333-0. [DOI] [PubMed] [Google Scholar]
- 18.Dabert P, Smith G R. Genetics. 1997;145:877–889. doi: 10.1093/genetics/145.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng A, Tang H, Ong B A, Farrell M J, Lin S. Proc Natl Acad Sci USA. 1997;94:6267–6272. doi: 10.1073/pnas.94.12.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cormack B P, Valdivia R H, Falkow S. Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 21.Vieira J, Messing J. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 22.Detrich H W, III, Kieran M W, Chan F Y, Barone L M, Yee K, Rundstadler J A, Pratt S, Ransom D, Zon L I. Proc Natl Acad Sci USA. 1995;92:10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casadaban M C, Cohen S N. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 24.Morrison D A. Methods Enzymol. 1979;68:326–331. doi: 10.1016/0076-6879(79)68023-0. [DOI] [PubMed] [Google Scholar]
- 25.Culp P, Nusslein-Volhard C, Hopkins N. Proc Natl Acad Sci USA. 1991;88:7953–7957. doi: 10.1073/pnas.88.18.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant S G N, Jessee J, Bloom F R, Hanahan D. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capecchi M R. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 28.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith G R. Experientia. 1994;50:234–241. doi: 10.1007/BF01924006. [DOI] [PubMed] [Google Scholar]
- 30.Myers R S, Stahl M M, Stahl F W. Genetics. 1995;141:805–812. doi: 10.1093/genetics/141.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen P, Huang H V. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long Q, Meng A, Wang H, Jessen J R, Farrell M J, Lin S. Development (Cambridge, UK) 1997;124:4105–4111. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- 33.Amsterdam A, Lin S, Hopkins N. Dev Biol. 1995;171:123–129. doi: 10.1006/dbio.1995.1265. [DOI] [PubMed] [Google Scholar]
- 34.Amsterdam A, Lin S, Moss L G, Hopkins N. Gene. 1996;173:99–103. doi: 10.1016/0378-1119(95)00719-9. [DOI] [PubMed] [Google Scholar]
- 35.Moss J B, Price A L, Raz E, Driever W, Rosenthal N. Gene. 1996;173:89–98. doi: 10.1016/0378-1119(95)00729-6. [DOI] [PubMed] [Google Scholar]
- 36.Wilson C, Bellen H J, Gehring W J. Annu Rev Cell Biol. 1990;6:679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
- 37.Driever W, Solnica-Krezel L, Schier A F, Neuhauss S C, Malicki J, Stemple D L, Stainier D Y, Zwartkruis F, Abdelilah S, Rangini Z, et al. Development (Cambridge, UK) 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 38.Haffter P, Granato M, Brand M, Mullins M C, Hammerschmidt M, Kane D A, Odenthal J, van Eeden F J, Jiang Y J, Heisenberg C P, et al. Development (Cambridge, UK) 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]