Abstract
The XIST gene, expressed only from the inactive X chromosome, is a critical component of X inactivation. Although apparently unnecessary for maintenance of inactivation, XIST expression is thought to be sufficient for inactivation of genes in cis even when XIST is located abnormally on another chromosome. This repression appears to involve the association of XIST RNA with the chromosome from which it is expressed. Reactivated genes on the inactive X chromosome, however, maintain expression in several somatic cell hybrid lines with stable expression of XIST. We describe here another example of an XIST-expressing human–hamster hybrid that lacks X-linked gene repression in which the human XIST gene present on an active X chromosome was reactivated by treatment with 5-aza-2′-deoxycytidine. These data raise the possibility that human XIST RNA does not function properly in human–rodent somatic cell hybrids. As part of our approach to address this question, we reactivated the XIST gene in normal male fibroblasts and then compared their patterns of XIST RNA localization by subcellular fractionation and in situ hybridization with those of hybrid cells. Although XIST RNA is nuclear in all cell types, we found that the in situ signals are much more diffuse in hybrids than in human cells. These data suggest that hybrids lack components needed for XIST localization and, presumably, XIST-mediated gene repression.
Keywords: azacytidine, X chromosome inactivation, methylation, transcription, fluorescence in situ hybridization
Stable expression of XIST is required on the inactive X chromosome for the establishment of mammalian X chromosome inactivation (reviewed in ref. 1). The role of XIST in the maintenance of repression has been questioned, however. Previous studies of inactive X chromosomes with XIST deletions indicate that XIST RNA is not necessary to maintain X inactivation (2, 3), presumably because other repressive systems, such as promoter methylation, histone deacetylation, and/or late replication, are maintaining inactivation. Our studies of human–hamster hybrids containing an inactive X chromosome with azacytidine-reactivated genes indicate that XIST expression is not sufficient to prevent reactivation or to reinitiate silencing of these genes (4). A similar conclusion was reached by other workers studying reactivation of X-linked genes in another cell hybrid system (5).
To examine this phenomenon further, we reactivated the silent XIST gene on the human active X chromosome in a human–hamster hybrid and in normal human male fibroblasts. The rationale for reactivation was based on the apparent regulation of XIST expression by 5′-CG-3′ dinucleotide methylation. This region is hypermethylated on the silent, active X allele and is hypomethylated on the expressed, inactive X allele in both human (6, 7) and murine (8–11) somatic tissues. A further indication that 5′ hypermethylation is important in Xist regulation is that the active X allele is expressed in somatic cells of male mice deficient in DNA methyltransferase (12, 13).
Repression by 5′-CG-3′ dinucleotide methylation commonly is found for X-inactivated genes, and reactivation of such genes is accomplished easily in somatic cell hybrids by treatment with 5-aza-2′-deoxycytidine (5aCdr) (14), but it is not possible in normal cells such as fibroblasts (15). XIST was reactivated by 5aCdr treatment not only in an active X hybrid but also in normal human fibroblasts, presumably because of the absence of X inactivation-associated repressive systems that prevent gene reactivation from the inactive X. Although XIST reactivation was generally transient in both cell types, a hybrid clone with stable expression was isolated and studied for expression of X-linked genes that are subject to X inactivation. No evidence of XIST-mediated repression was found; these results are similar to our studies of such genes that were reactivated in inactive X hybrids.
These results are surprising in view of the continued expression of XIST in all female somatic cells and the close association of XIST RNA to the inactive X. Reasoning that the absence of XIST-mediated gene repression in hybrids may be the result of abnormal function in such cells, we conducted subcellular fractionation and fluorescent in situ hybridization (FISH) analysis of XIST RNA in hybrids and human fibroblasts. Our findings suggest that proper localization of XIST RNA may be required for repressive activity and that species specificity may explain the abnormal localization patterns we found in hybrids.
MATERIALS AND METHODS
Cell Culture.
Standard growth conditions for cultured cells were as described (16). X8–6T2S1 is a human–hamster hybrid subclone that contains a single human inactive X chromosome, and Y162–11CS3 is a similarly derived hybrid that contains an active X chromosome (16). GM06318 (NIGMS Human Genetic Mutant Cell Repository) is a human fibroblast–hamster hybrid cell line that contains a single human chromosome that is an active X chromosome. The human–hamster hybrid 8121-TGRD contains an inactive human X chromosome that is deleted terminally distal to Xq26 (4, 17). III-5 and 19AS2 are inactive X hybrids that express XIST and contain azacytidine-reactivated X-linked genes (4). Hybrid lines were confirmed to contain a human X chromosome in >90% of cells by using standard cytogenetic banding methods. Normal human fibroblasts were obtained as frozen stocks from G. Martin (Department of Pathology, University of Washington) and grown in Amniomax-C100 medium (GIBCO/BRL). Reactivation was induced with 5aCdr treatment of cells in log phase growth (18).
Reverse Transcription (RT) PCR Analysis.
Total RNA was isolated, and RT-PCR analysis was performed as described by using 0.5 and 1.0 μg of RNA in the RT reaction (18). In some experiments, nuclear and cytoplasmic RNAs were prepared after the fractionation procedure described in Sambrook et al. (19) in which cells are lysed with nonionic detergent and nuclei are separated by centrifugation. Amplification of cDNA was performed as described by using the following primer sets: XIST, xst8:9r; PGK1, pgk1-R3:R4; MIC2, XMIC2:XMIC2R; and SLC16A2 (XPCT), xpct-A3.2:A5.2 (4). For semiquantitative analyses, amplification was limited to between 18 and 21 cycles so as to remain in the exponential range. A portion (15%) of the PCR product was subject to agarose gel electrophoresis and Southern blot analysis. To screen early stage clones of 5aCdr-treated GM06318, RNA was extracted from ≈5 × 105 cells and subjected to RT-PCR without prior quantification; products were analyzed by agarose gel electrophoresis and ethidium bromide staining. To quantify the level of reactivation in 5aCdr-treated male fibroblasts, 25 cycles of PCR were carried out for MIC2 and XIST, and 5% of the product was subject to electrophoresis, Southern analysis, and PhosphorImager (Molecular Dynamics) quantification.
DNA Methylation Analysis.
Genomic DNA was isolated from cultured cells, digested with restriction enzymes, and analyzed by Southern analysis as described (20). The reaction buffer for PstI and PstI–SacII double digests was 50 mM Tris⋅HCl (pH 8), 50 mM NaCl, 10 mM MgCl2, 0.1 mg/ml BSA, 1 mM DTT, 1 mM spermidine, 9 units of each restriction enzyme, and 3 μg of DNA. Membranes were hybridized with xst31:29r, a 530-bp, PCR-generated probe (21, 22).
FISH Analysis of XIST RNA.
Cell preparation. Cells were grown on alcohol-washed and UV-sterilized glass coverslips (22 × 22 mm) in 35-mm Petri dishes. After removal of the medium, cells were washed twice in PBS and fixed in three changes of 3:1 methanol:acetic acid (2 min, 2 min, and 15 min). Coverslips were air-dried and were used immediately for hybridization. Mass cultures of 5aCdr-treated human fibroblasts were examined between 2 and 14 days after treatment; mass cultures of the GM06318 hybrid were examined 5 days after 5aCdr treatment.
Probe preparation.
The XIST probe D6122 (23) is a gift from A. C. Chinault (Baylor College of Medicine, Houston, TX). The cosmid (5 μg) was biotinylated by using a BioNick kit obtained from GIBCO/BRL. Human COT-1 DNA (150 μg) (GIBCO/BRL) and sheared salmon testes DNA (50 μg) were added to the reaction product before ethanol precipitation. The mixture was dissolved in 100 μl of 50% formamide, 2 × standard saline citrate, and 10% dextran sulfate and stored at −20°C before use.
Hybridization and detection.
The biotinylated probe mixture (4 μl/coverslip) was denatured for 5 min at 70°C and preannealed at 37°C for 30–60 min. Probe (4 μl) containing 0.4 μl of vanadyl ribonucleoside complex (GIBCO/BRL) was placed on cells, covered with a plastic coverslip, and placed in a humidified chamber for 4 hr at 37°C. Procedures for washing and detection with Texas Red avidin were as described in the Oncor protocol for unique probes. The coverslip was mounted in 4, 6-diamido-2-phenylindole dihydrochloride (DAPI) antifade solution. Slides were examined with a Nikon Microphot-FXA microscope, and cells were photographed under epifluorescence by using a triple bandpass filter.
For dual XIST RNA and X-specific α-satellite DNA FISH analysis, the methods of Clemson et al. (24) were followed with two exceptions: (i) The original fixation was 3:1 methanol:acetic acid, followed by drying and hybridization with the XIST probe, and (ii) the paraformaldehyde fixation after XIST hybridization was carried out at an alkaline pH (pH 9–10). The digoxygenin-labeled X chromosome α-satellite probe (Oncor) was used according to the manufacturer’s instructions. Images of DAPI, Texas Red, and fluorescein isothiocyanate were collected sequentially without image-shift by using selective bandpass excitation filters in a computer-controlled filter wheel (Ludl, Hawthorne, NY) and a multiple bandpass emission filter (ChromaTechnology, Brattleboro, VT). Images were digitized by using a Princeton air-cooled charge-coupled device camera with a Kodak KAF1400 chip operated in the two-by-two binning mode. Digital images were processed by using IPLab Spectrum software v. 3.0 (Scanalytics, Billerica, MA). DAPI bands were sharpened by using the built-in “Hat” filtering process and were displayed as gray values. The threshold and contrast of the fluorescein isothiocyanate and Texas Red images were manipulated to facilitate identification of dim sites. The fluorescein isothiocyanate and Texas Red images then were pseudocolored in green and red, respectively, and overlaid on the DAPI image for analysis.
RESULTS
Reactivation of XIST in a Somatic Cell Hybrid.
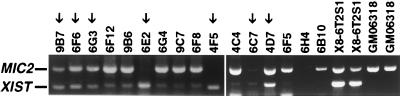
Reactivation of XIST on the human active X chromosome in the human–hamster hybrid, GM06318, occurred after treatment of exponentially growing cells with 5aCdr. Early stage clones (≈15 doublings) were screened for XIST and MIC2 expression by RT-PCR; PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining as shown in Fig. 1. XIST transcription was detected in the inactive X human–hamster hybrid X8–6T2S1 but not in the untreated active X hybrid GM06318. XIST reactivation was observed in a large proportion of 5aCdr-treated clones (20 of 48 clones). Clones with strong XIST expression were identified by comparing the relative extent of ethidium staining for the XIST and MIC2 RT-PCR products in reactivant clones with that in X8–6T2S1 cells. Several such clones were expanded and maintained in culture for several generations to determine the stability of XIST reactivation.
Figure 1.
RT-PCR analysis of XIST and MIC2 (escapes X inactivation) expression in clones derived from the active X hybrid, GM06318, after 5aCdr treatment. Unselected clones of 5aCdr-treated GM06318 were examined for XIST reactivation at an early stage of growth by RT-PCR. Representative data are shown from a total of 48 clones analyzed that were positive for expression of MIC2, XIST, or both. Some clones were positive for XIST and not MIC2 because of a spurious problem with MIC2 amplification in duplex PCR at low template concentrations (data not shown). Control cells included GM06318 and the inactive X hybrid X8–6T2S1. Arrows point to clones scored as positive for XIST reactivation.
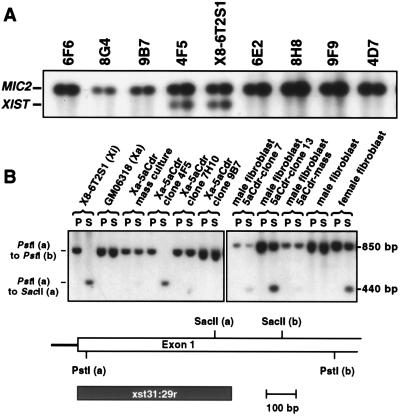
Semiquantitative RT-PCR analysis of XIST expression relative to that of MIC2 indicated that reactivation was mostly transitory; only 1 clone of 11 examined, named 4F5, exhibited stable expression during continued growth over several generations at a level similar to that found in the inactive X hybrid (Fig. 2A). In addition, subsequent subcloning analysis of 4F5 indicated that all seven subclones analyzed exhibited XIST levels similar to those in the parental culture (data not shown). Unmethylated SacII sites in the 5′ region of XIST are characteristic of the expressed allele (7), and we found that only the 4F5 clone and the inactive X hybrids show a pattern of complete SacII digestion in this region (Fig. 2B). The fully methylated revertant clones, such as 7H10 and 9B7, presumably were unmethylated at the earlier stage when XIST was being expressed.
Figure 2.
Stability of XIST reactivation in active X hybrid clones treated with 5aCdr and DNA methylation analysis. (A) XIST-positive clones derived from the 5aCdr-treated GM06318 culture were expanded and maintained for several generations to determine the stability of XIST reactivation. XIST expression was analyzed in these clones and X8–6T2S1 by semiquantitative RT-PCR as described in Materials and Methods; shown is a Southern blot analysis of XIST:MIC2 RT-PCR products. All samples were run at two concentrations in the RT reaction (0.5 or 1.0 μg RNA/reaction). (B) Methylation analysis of the 5′ region of XIST. Genomic DNA was digested with either PstI alone or with PstI and SacII together. SacII sites in the 5′ region of XIST are unmethylated on the expressed allele and methylated on the repressed allele in human cells or in cell hybrids. 4F5, 7H10, and 9B7 are 5aCdr-treated GM06318 clones that exhibited strong XIST reactivation at a very early stage of growth. Clones derived from 5aCdr-treated normal male fibroblasts were analyzed after growing to ≈3 × 106 cells.
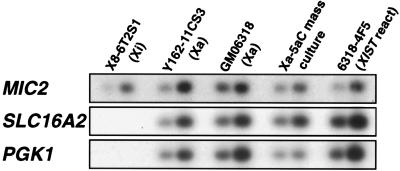
To investigate whether XIST expression in 4F5 silences genes subject to X inactivation, we examined the transcription of two such genes located near XIST, SLC16A2 (XPCT) and PGK1, by using semiquantitative RT-PCR with MIC2 expression as a normalizing standard. As seen in Fig. 3, expression of PGK1 and SLC16A2 in the 4F5 reactivant was similar to that in the untreated parental hybrid GM06318. Selection against inactivation of the human X-linked genes is not likely because hybrid cells should be able to survive and function without the human X under normal growth conditions, as the parental hamster cells and inactive X hybrids do.
Figure 3.
Semiquantitative RT-PCR analysis of gene expression for genes located near XIST in the inactive X hybrid X8–6T2S1, the active X hybrids Y162–11CS3 and GM06318, the uncloned mass culture of GM06318 after 5aCdr treatment, and the 4F5 XIST reactivant clone derived from GM06318. Southern blot analyses of PGK1 and SLC16A2 (XPCT) RT-PCR products are shown in relation to that of MIC2 products; the same RT products from each cell line (random-primed) were used as templates for all three gene-specific PCR primers. All samples were run at two concentrations in the RT reaction (0.5 or 1.0 μg RNA/reaction).
Nuclear Localization of XIST RNA.
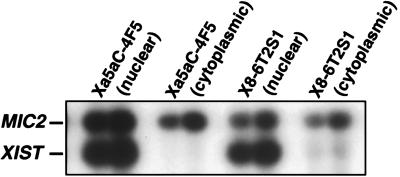
Because XIST RNA localizes to the nucleus in female somatic cells and is associated closely with the inactive X chromosome (12, 13, 21, 24), we wanted to determine whether the lack of XIST-induced repression of genes subject to X inactivation in the 4F5 reactivant might be explained by nonnuclear localization of XIST or improper processing of the transcript. We isolated RNA from nuclear and cytoplasmic extracts and found by RT-PCR that the spliced XIST RNA was localized properly to the nuclear fraction in the 4F5 cells (Fig. 4). In 5aCdr-treated inactive X hybrids reactivated for X-inactivated genes despite high levels of XIST expression (4), we also observed nuclear localization of spliced XIST (Fig. 4). Nuclear localization of XIST RNA, therefore, is not sufficient to promote X inactivation.
Figure 4.
Subcellular localization of XIST RNA. XIST RNA is known to localize to the nucleus in female somatic cells, and it apparently is associated with the inactive X chromosome. To determine whether XIST localization was normal in the inactive X hybrid (X8–6T2S1) and the XIST reactivant hybrid (Xa5aC-4F5), we isolated RNA from cytosolic and nuclear extracts and analyzed these fractions by semiquantitative RT-PCR. PCR products were derived from either 0.5 or 1.0 μg of RNA in the RT reaction (see Materials and Methods).
XIST Reactivation in Normal Male Fibroblasts.
Although reactivation of X-inactivated genes has not been reported in normal fibroblasts, we speculated that the repressed allele of XIST on the active X chromosome might be reactivated by demethylation of its promoter region after 5aCdr treatment because of its early replication in the cell cycle (22). Such replication timing should make it more permissive to transcription, and thus reactivation, than X-inactivated genes that are methylated and that replicate late in S phase (4).
Treatment of male fibroblasts with a single exposure to 5aCdr resulted in 23 of 73 clones that were positive for XIST, a rate comparable to that found after similar treatment of the active X hybrid. Reactivation was detectable after two cell doublings after 5aCdr treatment and was transitory. The level of expression, however, was much lower in the normal fibroblast than in the hybrid cells; expression levels among 5aCdr-treated clones harvested 5–10 doublings after treatment varied from <0.16% up to 3.4% of normal female levels among 21 clones examined. These levels were calculated by comparing the XIST/MIC2 ratio for treated clones with that of normal female cells. As in the 4F5 hybrid reactivant, XIST RNA in 5aCdr-treated male fibroblasts was confined to the nucleus (data not shown).
Although there was a low level of reactivation in 5aCdr-treated fibroblasts, some clones were found to be unmethylated at a 5′ SacII site in ≈50% of cells (Fig. 2B, clones 7 and 13). Other sites in the 5′ region were not examined, however, and these are likely to contribute to XIST repression if methylated. We found no evidence of reduced PGK1 expression in the cultures of 5aCdr-treated fibroblasts (data not shown), but such analyses are not likely to determine such repression in XIST reactivant cells because the low XIST expression levels we observed in mass cultures or clones apparently derive from a small number of cells (see next section).
RNA FISH Analysis of XIST in the 4F5 Reactivant, Inactive X Hybrids, and Normal Fibroblast Reactivants.
To determine whether the nuclear localization of XIST in the 4F5 reactivant and 5aCdr-treated male fibroblasts resembled the compact, inactive X-associated XIST structure that is present in normal human female cells (24), we examined these cells by using FISH analysis. Whereas human male cells lacked any XIST RNA signal (Fig. 5A), female fibroblasts consistently displayed a strong signal that was largely cohesive (Fig. 5B). That the FISH signal resulted from hybridization to XIST RNA was verified by its sensitivity to RNase H digestion (data not shown; see ref. 24). Signals routinely were detected in virtually all cells, and the majority of signals (>60%) were at the circumferential edge of the relatively flattened nuclei, a characteristic location for sex chromatin in fibroblasts. Hybrid cells containing an active X chromosome as the only human chromosome were consistently negative (data not shown).
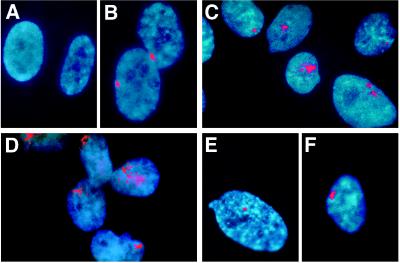
Figure 5.
FISH analysis of XIST RNA localization. To examine further the characteristics of the XIST RNA product in reactivant cells, its cellular location was examined by hybridization to a genomic cosmid clone containing XIST RNA sequences. Cells were fixed with 3:1 methanol to acetic acid and hybridized with biotin-labeled probe without prior denaturation of cellular DNA. (A) Normal human male fibroblasts. (B) Normal human female fibroblasts. (C) The XIST reactivant clone 4F5 derived from GM06318 after 5aCdr treatment. (D) The inactive X human–hamster hybrid 8121-TGRD. (E and F) XIST reactivant cells derived from a normal human male fibroblasts after 5aCdr treatment.
Inactive X hybrids, inactive X hybrids with reactivated genes, and the 4F5 active-X hybrid reactivated for XIST expression had much different XIST signals than those of female fibroblasts. The hybrid signals were more variable in appearance, often weaker than those in female fibroblasts; the strong hybrid signals were more diffuse than those seen in female fibroblasts. Furthermore, the signals in hybrid cells were not as frequently located at the edge of the nucleus as in female fibroblasts (Fig. 5 C and D; data not shown). We also examined XIST localization in uncloned cultures of the active X hybrid soon after 5aCdr treatment to estimate the initial reactivation frequency for XIST and to determine whether abnormal XIST RNA localization occurs before cloning and continued growth in culture. XIST signals were found in ≈10% of cells in these cultures, and they were quite similar to signals found in 4F5 cells and the inactive X hybrids (data not shown), suggesting that abnormal localization of XIST in hybrids cannot be attributed to selection during continued growth.
RNA FISH analysis of 5aCdr-treated human male fibroblast cultures were largely negative (1% or less positive). Clones derived from such cultures also contained only a few positive cells. Cells positive for XIST often had small, cohesive signals (Fig. 5E) and occasionally displayed larger signals resembling those found in female fibroblasts (Fig. 5F). The high frequency of negative cells in these cultures implies that the low overall level of XIST expression in 5aCdr-treated male fibroblasts ascertained by RT-PCR derives from a modest-to-normal level of expression in a very small percentage of cells.
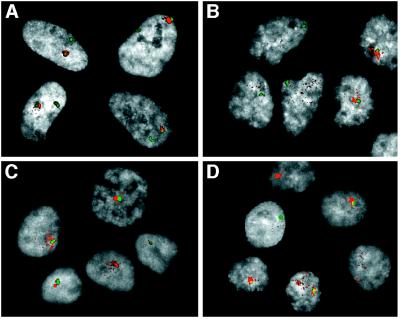
The diffuse nature of XIST signals in hybrid cells may result from a lack of association with the X chromosome. To examine this possibility, we performed dual labeling experiments for simultaneous detection of XIST RNA and X-chromosomal α-satellite sequences. The XIST signal in normal female cells always was associated with one of the two α-satellite signals, as expected (Fig. 6A). In such cells, we also observed a further association of the XIST signal with the DAPI-stained sex chromatin, thus confirming a close association with the inactive X chromosome (data not shown). Because many of the XIST signals in hybrid cells are weak, we selected optical fields with strong signals for image analysis. In such cells, much of the XIST signal was often completely separate from the α-satellite signal and often spanned a distance that was greater than that which separates the two α-satellite sequences in normal female cells (Fig. 6 B–D). In addition, the sex chromatin type of DAPI staining that coincides with XIST signals in normal female cells was not observed in hybrid cells.
Figure 6.
Dual-label FISH analysis of XIST RNA and X-specific α-satellite DNA. To examine further the disperse localization of XIST RNA in hybrid cells, the XIST signal (red) was detected as in Fig. 5, and the cells then were fixed in paraformaldehyde for subsequent denaturation and detection of X-specific α-satellite sequences (green). DAPI staining of nuclei is represented in gray scale (see Materials and Methods). (A) Normal female fibroblasts. (B) The inactive X human–hamster hybrid X8–6T2S1. (C) The inactive X human–hamster hybrid 8121-TGRD. (D) The XIST reactivant hybrid clone 4F5.
DISCUSSION
Reactivation of XIST in a Somatic Cell Hybrids and Normal Human Fibroblasts.
Based on studies suggesting a permissive effect of early replication on transcriptional activity (25–27), we hypothesized that the early replication of the silent, hypermethylated XIST allele should allow it to be reactivated readily by demethylation of the promoter region. We found that XIST reactivation occurred with high frequency after 5aCdr treatment of both an active X hybrid cell line and normal male fibroblasts. These high rates of reactivation are quite unusual, particularly in the normal human fibroblasts, because X-inactivated genes fail to be reactivated by 5aCdr treatment in these cells (15). Although reactivation of XIST in both the somatic cell hybrid and the male fibroblasts was generally transient, one stable hybrid reactivant was isolated. Based on our limited methylation analysis (Fig. 2), this instability likely involves remethylation of the 5′ region of XIST.
The level of XIST RNA in the hybrid reactivant was similar to that in hybrids with an inactive human X chromosome and was localized to the nuclei of both the hybrid and the normal fibroblast reactivants. Inactivation of X-linked genes, however, was not detected in these XIST reactivants. RNA FISH analysis of XIST localization revealed a distinct difference between female fibroblasts and hybrids containing either an inactive X chromosome or an active X with reactivated XIST. Although the XIST RNA signals in female fibroblasts resembled typical sex chromatin morphology, those in hybrid cells differed; many hybrid signals appeared weak, and strong ones were usually diffuse (Figs. 5 and 6). These patterns suggest that the failure of XIST RNA to cause or maintain gene repression in the hybrids results from an impaired association with the X chromosome or that such an association, if it occurs, is insufficient to promote an inactive X-like chromatin condensation.
Although reactivation of XIST in normal male fibroblasts occurred with high frequency, only a small percentage of cells in a mass culture or in reactivated clones were positive for XIST RNA. FISH signals for XIST RNA in these cells, however, appeared more like the normal signal found in female fibroblasts than the diffuse signals found in hybrids (Fig. 5). We speculate that two factors could account for the very low percentage of cells expressing XIST in these 5aCdr-treated male fibroblasts: (i) Remethylation of 5aCdr-induced demethylation occurs with greater efficiency than in treated hybrids, and/or (ii) cells that do express XIST become terminal or nonreplicative because of the resulting silencing of the active X chromosome. The observation that most cells lack XIST signals, even in clones of 5aCdr-treated male fibroblasts, is consistent with the possibility that many cells have reverted to XIST repression because of remethylation. This explanation is supported by our finding that, in the two fibroblast clones examined, ≈50% of the cells in both clones were methylated at a SacII site in the 5′ region (Fig. 2B). In the cells that were unmethylated at this site, remethylation may have occurred at other sites important for XIST repression.
The continued presence of XIST-positive cells in the male fibroblast population suggests that XIST-mediated lethality does not occur rapidly in these cells (1–2 weeks in culture). An alternative explanation is that XIST expression may switch between “on” and “off” states in the population such that those cells surviving by XIST repression have a tendency to switch back to the on state at a low rate, thus leading to XIST-mediated cell death for a small percentage of the population. Unfortunately, the low frequency of transmission for the reactivated state and the low number of expressing cells made this system a difficult one to analyze experimentally.
The striking difference between the ease of reactivation in human–rodent hybrids with an inactive human X chromosome and the difficulty of reactivation in normal fibroblasts led to the idea that a multiplicity of different repressive factors operate in normal cells to maintain the inactive X in a repressed state (28). The permissiveness of hybrid cells to reactivation was assumed to result from a compromised repressive system. At least four features are known to distinguish the active from the inactive X chromosome: promoter methylation, replication timing, XIST expression/XIST localization, and histone acetylation patterns; these systems should provide a strong hindrance to reactivation if they act independently to repress transcription.
In the case of XIST on the active X chromosome, our observations of reactivation might be explained by at least two of these repressive systems being absent: late replication timing and XIST RNA-mediated silencing. Holliday and Ho (29) reported 5aCdr-induced reactivation of HPRT in a human male fibroblast. The gene had been inactivated after electroporation of 5-methyl dCTP and likely was to be in a permissive state for reactivation because of the absence of XIST-mediated silencing (no XIST expression) and the probable retention of its early replication after 5-methyl dCTP treatment.
The absence of XIST-mediated silencing and other X inactivation systems might also explain the recently reported 5aCdr-induced reactivation of FMR1 on active X chromosomes of fragile X lymphoblasts (30). Reactivation of imprinted genes by using 5aCdr also has been examined. Hu et al. (31) described 5aCdr-induced reactivation in normal brain astrocytes of the repressed allele of an imprinted autosomal gene, IGF2. In addition, reactivation of the repressed H19 allele was observed in rabdomyosarcoma cells after 5aCdr treatment (32). The repressed alleles at imprinted loci often are characterized by promoter methylation and sometimes by late replication. We found that replication of the repressed alleles of IGF2 and H19 in normal fibroblasts occurs in mid-S phase (33), the same time as that of the expressed alleles. According to our hypothesis, the finding of frequent reactivation of XIST by 5aCdr should also apply to IGF2 and H19 because, in each case, replication timing is permissive for transcription.
Absence of XIST-Mediated Gene Repression and Abnormal Localization of XIST RNA in Somatic Cell Hybrids.
The idea that XIST RNA is not necessary to maintain X inactivation is derived from reports that inactivation is retained in cells with XIST deletions (2, 3). The question, however, of whether XIST expression is sufficient to maintain X inactivation in somatic cells is not answered by those observations; inactivation likely is maintained in these cells by redundant repressive mechanisms. Our finding of 5aCdr reactivation of several X-inactivated genes in human–hamster hybrids expressing XIST indicates that XIST expression is not sufficient for the maintenance of inactivation (4). Yoshida et al. (5) reached a similar conclusion for X-linked genes reactivated in a human–mouse hybrid system. This conclusion also follows from our studies of the 4F5 XIST reactivant reported here, although our observations of abnormal XIST RNA localization in 4F5 and the inactive X hybrids suggest that XIST does not function normally in hybrid cells.
It seems likely that XIST RNA, when functioning normally, is sufficient for the maintenance of X inactivation in somatic cells in the absence of methylation. This hypothesis is supported by the early observations of Kratzer et al. (34) that X inactivation of genes in murine extraembryonic cells (now known to express XIST) does not involve modification of their DNA. In transgenic mice and embryonic stem cells, the incorporation of tandem copies of Xist transgenes with as little as 15 kilobases of flanking sequence appears to be sufficient for repression of linked genes (35, 36). In transgenic mice deficient in DNA methyltransferase, activation of the endogenous Xist in male cells appears to result in the repression of the closely linked Pgk1 gene (13). The latter result argues against a requirement of some sort of interaction between Xist alleles for silencing as might be imagined in the Xist transgenic mice. Given the differences between our experiments and these transgenic data, one explanation of our observation that XIST does not repress genes in somatic cell hybrids might be that additional processes or factors are required for silencing that are only present during early development.
The apparent failure of XIST RNA to localize normally in human–rodent hybrids implies yet another factor in the X-inactivation system. That the hamster genome is dominant in human–hamster hybrids is evident from the fact that a full complement of hamster chromosomes is retained, whereas many or most human chromosomes are lost. A crucial XIST-interacting molecule or molecules normally may be expressed in somatic cells, but in hybrids, these factors may be only of hamster origin and may not be recognized properly by human XIST RNA, which diverges ≈30% from the murine sequence (21, 37). This idea is consistent with the apparently normal XIST RNA patterns found in 5aCdr-treated human male fibroblasts. An alternative explanation is that the human and/or hamster homologues of such genes could be present in the hybrids but in a repressed state (38–40).
It has been reported that the up-regulation of Xist expression that occurs in association with murine X chromosome inactivation results from stabilization of the Xist transcript on the inactive X (41, 42). Before X inactivation, XIST is transcribed from both X chromosomes at low levels (41–43), and this expression can be visualized by RNA FISH as two small dots corresponding to sites of transcription on each X (41, 42). Our studies indicate that XIST RNA stability is not the only determinant of normal XIST localization because abnormal RNA FISH signals were seen in hybrids, even though the level of expression is similar to that of normal female cells when analyzed by RT-PCR (ref. 22 and Fig. 4). Consistent with this high level of expression, the XIST RNA FISH signals in hybrids, although diffuse, are much larger than the “site-of-transcription” signals that typify the unstable transcripts identified before X inactivation in the murine studies.
The data presented here further support a major role for promoter methylation in the regulation of XIST transcription and indicate that such transcription is not sufficient for normal chromosomal localization or for repression of cis-linked genes that are subject to X inactivation. Our XIST reactivant and the inactive X somatic cell hybrids should be useful in identifying factors that promote normal XIST localization and function.
Acknowledgments
We thank Hillary Massa and Barbara Trask for help with charge-coupled device analysis of dual-label FISH experiments, Craig Chinault for supplying the D6122 XIST cosmid probe, Carolyn Brown for helpful discussions, and Lester Goldstein for excellent help with manuscript revisions. This work was supported by grants from the National Institutes of Health (HD16659 and GM52463).
ABBREVIATIONS
- 5aCdr
5-aza-2′-deoxycytidine
- FISH
fluorescence in situ hybridization
- DAPI
4, 6-diamido-2-phenylindole dihydrochloride
- RT
reverse transcription
References
- 1.Lee J T, Jaenisch R. Curr Opin Genet Dev. 1997;7:274–280. doi: 10.1016/s0959-437x(97)80138-4. [DOI] [PubMed] [Google Scholar]
- 2.Brown C J, Willard H F. Nature (London) 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- 3.Rack K A, Chelly J, Gibbons R J, Rider S, Benjamin D, Lafreniere R G, Oscier D, Hendriks R W, Craig I W, Willard H F, et al. Hum Mol Genet. 1994;3:1053–1059. doi: 10.1093/hmg/3.7.1053. [DOI] [PubMed] [Google Scholar]
- 4.Hansen R S, Canfield T K, Fjeld A D, Gartler S M. Hum Mol Genet. 1996;5:1345–1353. doi: 10.1093/hmg/5.9.1345. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida I, Nishita Y, Mohandas T K, Takagi N. Exp Cell Res. 1997;230:208–219. doi: 10.1006/excr.1996.3393. [DOI] [PubMed] [Google Scholar]
- 6.Lafreniere R G, Willard H F. Genomics. 1993;17:502–506. doi: 10.1006/geno.1993.1356. [DOI] [PubMed] [Google Scholar]
- 7.Hendrich B D, Brown C J, Willard H F. Hum Mol Genet. 1993;2:663–672. doi: 10.1093/hmg/2.6.663. [DOI] [PubMed] [Google Scholar]
- 8.Heard E, Simmler M C, Larin Z, Rougeulle C, Courtier B, Lehrach H, Avner P. Genomics. 1993;15:559–569. doi: 10.1006/geno.1993.1108. [DOI] [PubMed] [Google Scholar]
- 9.Ariel M, Robinson E, McCarrey J R, Cedar H. Nat Genet. 1995;9:312–315. doi: 10.1038/ng0395-312. [DOI] [PubMed] [Google Scholar]
- 10.Norris D P, Patel D, Kay G F, Penny G D, Brockdorff N, Sheardown S A, Rastan S. Cell. 1994;77:41–51. doi: 10.1016/0092-8674(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 11.Zuccotti M, Monk M. Nat Genet. 1995;9:316–320. doi: 10.1038/ng0395-316. [DOI] [PubMed] [Google Scholar]
- 12.Beard C, Li E, Jaenisch R. Genes Dev. 1995;9:2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- 13.Panning B, Jaenisch R. Genes Dev. 1996;10:1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- 14.Gartler S M, Goldman M A. Dev Genet (Amsterdam) 1994;15:504–514. doi: 10.1002/dvg.1020150609. [DOI] [PubMed] [Google Scholar]
- 15.Wolf S F, Migeon B R. Nature (London) 1982;295:667–671. doi: 10.1038/295667a0. [DOI] [PubMed] [Google Scholar]
- 16.Hansen R S, Ellis N A, Gartler S M. Mol Cell Biol. 1988;8:4692–4699. doi: 10.1128/mcb.8.11.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ledbetter S A, Schwartz C E, Davies K E, Ledbetter D H. Am J Med Genet. 1991;38:418–420. doi: 10.1002/ajmg.1320380254. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki T, Hansen R S, Gartler S M. Mol Cell Biol. 1992;12:3819–3826. doi: 10.1128/mcb.12.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 7.12–7.13. [Google Scholar]
- 20.Hansen R S, Gartler S M. Proc Natl Acad Sci USA. 1990;87:4174–4178. doi: 10.1073/pnas.87.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown C J, Hendrich B D, Rupert J L, Lafreniere R G, Xing Y, Lawrence J, Willard H F. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 22.Hansen R S, Canfield T K, Gartler S M. Hum Mol Genet. 1995;4:813–820. doi: 10.1093/hmg/4.5.813. [DOI] [PubMed] [Google Scholar]
- 23.Boggs B A, Chinault A C. Proc Natl Acad Sci USA. 1994;91:6083–6087. doi: 10.1073/pnas.91.13.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clemson C M, McNeil J A, Willard H F, Lawrence J B. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown E H, Iqbal M A, Stuart S, Hatton K S, Valinsky J, Schildkraut C L. Mol Cell Biol. 1987;7:450–457. doi: 10.1128/mcb.7.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatton K S, Dhar V, Brown E H, Iqbal M A, Stuart S, Didamo V T, Schildkraut C L. Mol Cell Biol. 1988;8:2149–2158. doi: 10.1128/mcb.8.5.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatton K S, Schildkraut C L. Mol Cell Biol. 1990;10:4314–4323. doi: 10.1128/mcb.10.8.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gartler S M, Dyer K A, Graves J A, Rocchi M. Prog Clin Biol Res. 1985;198:223–235. [PubMed] [Google Scholar]
- 29.Holliday R, Ho T. Somatic Cell Mol Genet. 1995;21:215–218. doi: 10.1007/BF02254772. [DOI] [PubMed] [Google Scholar]
- 30.Chiurazzi P, Pomponi M G, Willemsen R, Oostra B A, Neri G. Hum Mol Genet. 1998;7:109–113. doi: 10.1093/hmg/7.1.109. [DOI] [PubMed] [Google Scholar]
- 31.Hu J F, Vu T H, Hoffman A R. J Biol Chem. 1996;271:18253–18262. doi: 10.1074/jbc.271.30.18253. [DOI] [PubMed] [Google Scholar]
- 32.Chung W Y, Yuan L, Feng L, Hensle T, Tycko B. Hum Mol Genet. 1996;5:1101–1108. doi: 10.1093/hmg/5.8.1101. [DOI] [PubMed] [Google Scholar]
- 33.Kawame H, Gartler S M, Hansen R S. Hum Mol Genet. 1995;4:2287–2293. doi: 10.1093/hmg/4.12.2287. [DOI] [PubMed] [Google Scholar]
- 34.Kratzer P G, Chapman V M, Lambert H, Evans R E, Liskay R M. Cell. 1983;33:37–42. doi: 10.1016/0092-8674(83)90332-x. [DOI] [PubMed] [Google Scholar]
- 35.Lee J T, Jaenisch R. Nature (London) 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 36.Herzing L B, Romer J T, Horn J M, Ashworth A. Nature (London) 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- 37.Brockdorff N, Ashworth A, Kay G F, McCabe V M, Norris D P, Cooper P J, Swift S, Rastan S. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 38.Miller D A, Dev V G, Tantravahi R, Miller O J. Exp Cell Res. 1976;101:235–243. doi: 10.1016/0014-4827(76)90373-6. [DOI] [PubMed] [Google Scholar]
- 39.Ajiro K, Zweidler A, Borun T, Croce C M. Proc Natl Acad Sci USA. 1978;75:5599–5603. doi: 10.1073/pnas.75.11.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hohmann P, Hohmann L K, Shows T B. Somatic Cell Genet. 1980;6:653–665. doi: 10.1007/BF01538644. [DOI] [PubMed] [Google Scholar]
- 41.Sheardown S A, Duthie S M, Johnston C M, Newall A E, Formstone E J, Arkell R M, Nesterova T B, Alghisi G C, Rastan S, Brockdorff N. Cell. 1997;91:99–107. doi: 10.1016/s0092-8674(01)80012-x. [DOI] [PubMed] [Google Scholar]
- 42.Panning B, Dausman J, Jaenisch R. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- 43.Tai H H, Gordon J, McBurney M W. Somatic Cell Mol Genet. 1994;20:171–182. doi: 10.1007/BF02254758. [DOI] [PubMed] [Google Scholar]