Abstract
Nodulation and nitrogen fixation genes of Mesorhizobium loti are encoded on the chromosome of the bacterium. Nevertheless, there is strong evidence that these genes can be transferred from an inoculant strain to nonsymbiotic mesorhizobia in the field environment. Here we report that the chromosomal symbiotic element of M. loti strain ICMP3153 is transmissible in laboratory matings to at least three genomic species of nonsymbiotic mesorhizobia. The element is 500 kb in size, integrates into a phe-tRNA gene, and encodes an integrase of the phage P4 family just within its left end. The entire phe-tRNA gene is reconstructed at the left end of the element upon integration, whereas the 3′ 17 nucleotides of the tRNA gene are present as a direct repeat at the right end. We termed the element a symbiosis island on the basis of its many similarities to pathogenicity islands. It may represent a class of genetic element that contributes to microbial evolution by acquisition.
The considerable impact of horizontal gene transfer on microbial evolution has become apparent from recent molecular analyses (1–4). For example, many bacterial pathogens contain clusters of virulence genes not present in closely related nonpathogenic strains or species. These gene clusters may be located on transmissible phage or plasmids, but are often found as so-called pathogenicity islands on the chromosome (5–8). The acquisition of a pathogenicity island is likely to have been a key step in the evolution of the pathogen. For example, the SPI-1 pathogenicity island was probably acquired very early in the evolution of Salmonella and enabled that genus to invade epithelial cells (9, 10). Pathogenicity islands range up to 190 kb in size, and most are found adjacent to or integrated within tRNA genes or flanked by insertion sequences. tRNA genes are targets for integration of plasmids and phage in a wide variety of bacteria, and pathogenicity islands in Dichelobacter nodosus and Vibrio cholerae are linked to phage-derived integrase genes, suggesting that pathogenicity islands may have been acquired by a phage-mediated process (11–13). However, transmissibility of pathogenicity islands has not been demonstrated and their evolutionary origin remains unknown.
We are working with Mesorhizobium loti, a species previously named Rhizobium loti, that is able to form nodules on several Lotus species. Mesorhizobium was recently described as a new genus of root nodule bacteria phylogenetically distinct from other root nodule bacteria of the genera Rhizobium and Bradyrhizobium on the basis of 16S rRNA gene sequence and other properties (14). M. loti differs from Rhizobium species and some other Mesorhizobium species in that its symbiotic information is chromosome rather than plasmid encoded (15–17). Nevertheless, we observed that genetically diverse symbiotic mesorhizobia, isolated from nodules off a stand of Lotus corniculatus established with a single inoculant strain, M. loti strain ICMP3153, in an area devoid of naturalized rhizobia able to nodulate the plant, contained an identical symbiotic DNA region to ICMP3153 (17). It was proposed that the genetically diverse symbionts arose through horizontal transfer of a chromosomal symbiotic element from the inoculant strain of M. loti to nonsymbiotic mesorhizobia (17). In support of this hypothesis, seven strains of nonsymbiotic mesorhizobia that belonged to four genomic species were subsequently isolated from the site where the diverse symbionts were found (18). The nonsymbionts were all auxotrophic for thiamin and biotin and all but one were auxotrophic for nicotinate (18). In contrast, ICMP3153 and the diverse symbiotic strains were prototrophic for all three vitamins, suggesting that the transferred element encodes genes required for vitamin synthesis as well as nodulation and nitrogen fixation genes.
Here we report transfer of the symbiotic element to three of the nonsymbiotic species, characterization of the ends and site of integration of the element, and determination of its size. The results indicate that nonsymbionts evolve into symbionts in a single step by acquisition of a 500-kb element that contains a phage-related integrase gene near one end and integrates into a tRNA gene. We have termed the transmissible element a symbiosis island in analogy to the pathogenicity islands of Gram-negative bacterial pathogens.
MATERIALS AND METHODS
Bacterial Strains.
Mesorhizobium strain ICMP3153 is the symbiotic inoculant strain used by Sullivan et al. (17). Strains CJ2, CJ3, CJ4, and CJ7 are nonsymbiotic Mesorhizobium strains that represent three genomic species (18). Strains R8CS, R8CL, R12C, R16C, and R88B are diverse symbiotic strains isolated by Sullivan et al. (17) and belong to the same genomic species as strains CJ3 and CJ4. ICMP3153 belongs to a fourth species (18).
Crosses.
Double mutants of the nonsymbiotic strains resistant to streptomycin and rifampicin were isolated for use as recipients in matings with ICMP3153 as donor. Cultures were grown in TY broth (18) at 28°C for 48 h, and 500 μl aliquots of donor and recipient cultures were mixed and filtered onto 2.5 cm diameter 0.45-μm nitrocellulose filters. Filters were incubated on TY plates for 24 h at 28°C. Cells were then washed off the filters and serially diluted onto plates of selective medium. Selective medium was G/RDM (19) without vitamins supplemented with streptomycin and rifampicin.
Cosmid Walking.
Two cosmid libraries of ICMP3153 DNA, one of partial EcoRI fragments cloned into pLAFR1 and the other of partial Sau3A fragments cloned into pIJ3200, were constructed as described (20, 21). End fragments of cosmids were subcloned, usually as EcoRI/PstI fragments, for use as probes for walking. Each cosmid identified in the walk was hybridized to Southern blots of EcoRI and SpeI digests of ICMP3153, nonsymbiotic strains, and their symbiotic derivatives to exclude chimeric cosmids and false steps associated with repeated regions.
The SpeI map was constructed by cosmid walking from a number of points within the symbiotic element by using the two cosmid libraries. p637 and p806 were identified as described below. Other probes used to initiate walking were p384 and p692. p384 contained a HindIII fragment fortuitously identified during selection of randomly cloned HindIII genomic fragments for use as restriction fragment length polymorphism probes (17). It hybridized to a 30-kb SpeI fragment. p692 contained an EcoRI/SpeI end fragment subcloned from the 120-kb SpeI fragment after excising the fragment from a pulse field gel. The 120-kb SpeI fragment was identified because it hybridized to pRLnodAC (17).
Other DNA Manipulations.
Procedures for genomic DNA isolation, pulse-field electrophoresis, and Southern hybridizations have been described (17). To amplify right junction fragments of the symbiotic element, genomic DNA from diverse symbiotic strains was digested with SalI plus XhoI (strains R8CS and R8CL) or with BspEI (strain R88B), ligated and used as template for inverse PCR. Inverse PCR was carried out with primers IP1 (5′-ACCGATCGAGTTCGTGGGCA) and IP2 (5′-GGTGCAGGTATGCCGGCGTCGT) that prime in opposite directions from within the right end of the element. Primers LE1 (5′-GAGCCCGCCACCGTCAGACA) and LE2 (5′-GTGCTATAACCCACGCGCTT) were designed from an alignment of the left junction sequences of ICMP3153, CJ4Sym, and CJ7Sym and used to amplify by PCR and sequence the left junction region from strains R8CS, R8CL, and R88B. This amplification product was also used as the phe-tRNA hybridization probe. For sequence analysis, PCR products were purified by using a QIAquick kit (Qiagen, Chatsworth, CA) and plasmid templates were prepared by using a Quantum plasmid miniprep kit (Bio-Rad). Sequences were determined on both strands by using combinations of subcloned fragments and custom primers and an Applied Biosystems model 373A autosequencer. Databases at the National Center for Biotechnology Information were searched by using blastn and blastx (22) for similar sequences via e-mail.
RESULTS
Transfer of the Symbiotic Element.
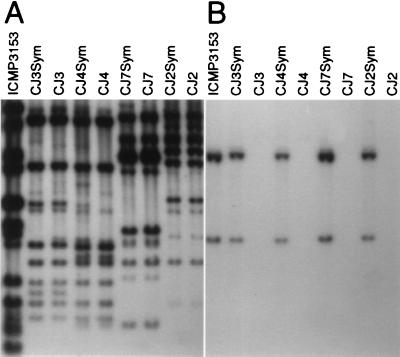
Strains CJ3, CJ4, and CJ7, representatives of three of the four nonsymbiont species isolated by Sullivan et al. (18), were auxotrophic for biotin, thiamin, and nicotinate, whereas the symbionts were prototrophic. These phenotypic differences were used as the basis of a selection system to detect transfer of the symbiotic element. Derivatives of CJ3, CJ4, and CJ7 that had gained vitamin prototrophy were selected following overnight incubation of the nonsymbionts with ICMP3153 on filters on agar plates. Colonies appeared after 5–8 days incubation at 28°C at a frequency of about 5 × 10−7 per recipient. These were purified and found to nodulate L. corniculatus when assayed for symbiotic ability on aseptically grown seedlings (18). Transfer of the element to the isolates was confirmed by Southern blot analysis (Fig. 1).
Figure 1.
Southern blot analysis demonstrating transfer of the symbiotic element to three species of nonsymbiotic mesorhizobia. Genomic DNA from the donor strain ICMP3153 and pairings of nonsymbiotic and symbiotic strains was digested with EcoRI and hybridized (A) with three random genomic HindIII fragments of ICMP3153 (18) and (B) with pRLnodAC containing M. loti nodAC genes (18).
The Symbiotic Element Integrates into a phe-tRNA Gene.
Two nonsymbiont/symbiont pairings (CJ4 and CJ4Sym; CJ7 and CJ7Sym) were used, together with inoculant strain ICMP3153 and three diverse symbiotic rhizobia that acquired the element in the field, to determine the site of integration of the element. Cosmid walking was used to identify a cosmid that spanned a junction, arbitrarily assigned the right junction, of the element in ICMP3153. The walk was initiated by using p637, a cosmid initially isolated because it complemented nonsymbionts to biotin and nicotinate prototrophy (unpublished data). Southern blot analysis of EcoRI digests of genomic DNA showed that p637 was fully conserved in diverse symbiotic strains and did not hybridize to nonsymbiotic strains, indicating it was part of the symbiotic element. When probed to Southern blots of SpeI digests of the same strains separated on pulse field gels, p637 hybridized to two bands in each symbiotic strain, a conserved band of 145 kb and a band unique to each strain, the smallest of which was 55 kb. This finding indicated p637 contained an SpeI site and mapped within 55 kb of one end of the integrated element.
End fragments of p637 were cloned, and the end clone that hybridized to the unique SpeI band was used to initiate cosmid walking. The fifth cosmid in the overlapping series, p778, hybridized to fragments conserved between all symbiotic strains and also to fragments not conserved with ICMP3153 but conserved between each member of the paired symbiotic and nonsymbiotic strains. Hence p778 contained the junction fragment which was subsequently subcloned as a 3.5-kb EcoRV fragment. This subclone did not hybridize to nonsymbiotic strains, indicating DNA immediately downstream of the element was not strongly conserved between the strains, but hybridized to a 1.8-kb PvuII junction fragment of CJ4Sym and a 1.1-kb SalI junction fragment of CJ7Sym. These fragments were cloned and found to hybridize to 1.3-kb EcoRI/PvuII and 1.1-kb SalI fragments from their respective nonsymbiotic counterparts, which were also cloned.
The cloned fragment from the nonsymbiont CJ4 that contained the target site of integration was used to identify fragments containing the left junction of the element in three symbionts. Hybridization with the 1.3-kb EcoRI/PvuII clone from CJ4 identified a 1.2-kb SalI left-junction fragment of CJ4Sym and a 1.3-kb left-junction PstI fragment of CJ7Sym that were cloned. The clone was also used to locate cosmid p806 from the ICMP3153 pLAFR1 library. p806 hybridized to fragments conserved between all symbiotic strains, to fragments not conserved with ICMP3153 but conserved between each pair of symbiotic and nonsymbiotic strains, and to one fragment unique to each strain. The unique fragments presumably spanned the left junction of the element in the symbionts and contained the integration target site in the nonsymbionts. A 6.1-kb EcoRI fragment containing the left junction region was subcloned from p806 for sequencing. The various junction and insertion site fragments were sequenced and primers were then designed from the sequences to amplify left and right junction fragments from the diverse symbiotic field isolates R8CS, R8CL, and R88B. These amplification products were also sequenced.
Comparative analysis of the sequences showed that the element inserted into a phe-tRNA gene in all strains (Fig. 2). The entire phe-tRNA gene was reconstructed at the left end of the element (Fig. 2B), whereas the 3′ 17 nucleotides of the gene were present as a direct repeat at the right end (Fig. 2C). The strains diverged immediately beyond the 17-bp region and thereafter fell into three homology groups that showed little similarity to each other in the 130-bp region sequenced for all strains (Fig. 2C and data not shown). In comparison, the sequence upstream of the tRNA was strongly conserved for at least 60 bp between the strains (Fig. 2B and data not shown).
Figure 2.
(A) Alignment of sequences of two nonsymbiotic strains CJ4 (AF049244) and CJ7 (AF049245) at the site of insertion. The phe-tRNA gene is marked in yellow and homologous flanking sequences are marked in blue. A phe-tRNA sequence from Neisseria gonorrhoeae (U82700) is included for comparison. (B) Alignment of sequences of six symbiotic strains at the left junction. Identical nucleotides upstream of the element are marked in blue, the phe-tRNA gene is marked in yellow, and the adjoining element DNA is in brown. (C) Alignment of sequences of six symbiotic strains at the right junction. Element sequence is marked in brown and the 17 nucleotides of the 3′ end of the phe-tRNA are marked in yellow. The three homology groups beyond the junction are indicated by like colors.
The Element Is at Least 500 kb in Size.
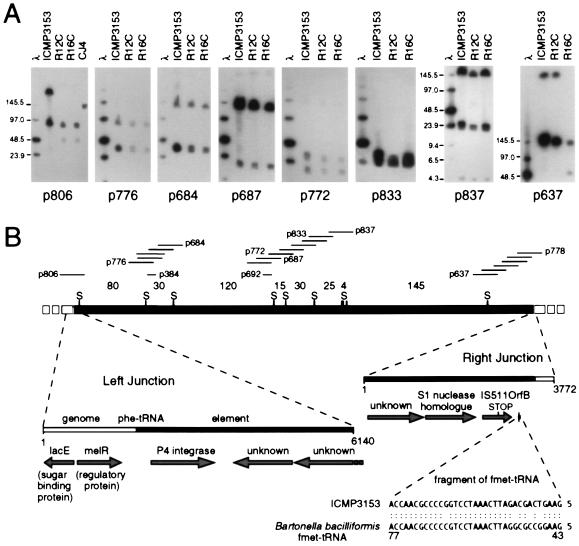
To determine the size of the integrated element, a provisional SpeI physical map of the element was constructed by cosmid walking from a number of points within the element (Fig. 3A). All SpeI fragments were linked but three large fragments within the element (80, 120, and 145 kb) were not fully covered (Fig. 3B). Hence the possibility of contiguous fragments of identical size was not eliminated. However the data establish that the element is at least 500 kb in size (Fig. 3B).
Figure 3.
(A) Linking of SpeI fragments of symbiotic element DNA by hybridization. R12C and R16C are diverse symbiotic strains that received the element from ICMP3153 in the field (17). Cosmids used for linking the fragments are named below the panels and are indicated together with other cosmids used for walking above the map in B. (B) Physical map of the element and location of features identified by sequencing of ICMP3153 DNA. Sizes of SpeI (S) fragments are shown in kilobases. There is 1.6 kb between the left junction of the element and the first SpeI site and 52 kb between the last SpeI site and the right junction. The sizes of the sequenced regions from ICMP3153 at the left junction (GenBank accession no. AF049242) and right junction (AF049243) are shown in base pairs. The region upstream of the element showed homology to the Escherichia coli melibiose regulatory gene melR (U14003), and the sugar-binding protein gene lacE from Agrobacterium radiobacter (X66596). The sequence at the right end homologous to fmet-tRNA is shown in comparison to sequence from Bartonella bacilliformis (L10238).
Nucleotide Sequences Within the Left and Right Ends of the Element.
Sequences of 6.1 kb at the left-end junction, including 4.2 kb within the element, and 3.8 kb at the right-end junction, including 3.4 kb within the element, were determined for strain ICMP3153. Within the left end of the element, an ORF of 419 aa initiated 198 bp downstream of the tRNA gene (Fig. 3B). This ORF was homologous (about 36% identity and 53% similarity over the full length) to members of the phage P4 integrase subfamily including retronphage phiR73 integrase and the integrases associated with pathogenicity islands in V. cholerae and D. nodosus (Fig. 4) (12, 13, 23–26). Two further ORFs of 385 aa and 391 aa (initiation codon beyond sequenced region) downstream of the integrase had no homologues in the databases. Upstream of the element were two divergently transcribed ORFs, a partial ORF with homology to sugar-binding proteins and a 290 aa ORF with homology (26% identity, 48% similarity) to a regulatory protein, MelR, involved in melibiose uptake in Escherichia coli (Fig. 3B).
Figure 4.
Alignment of the predicted amino acid sequence of the ICMP3153 symbiotic element integrase (3153 int) with sequences of some other members of the P4 integrase family: φR73 int, retronphage phi-R73 integrase (A42465); φSF6 int, Shigella flexneri phage Sf6 integrase (X59553); SlpA int, SlpA integrase from cryptic E. coli phage CP-57 (U36840); V. chol int, tcp-associated integrase from V. cholerae (U39068); D. nodo int, vap-associated integrase IntA from Dichelobacter nodosus (L31763). Identical amino acid residues are shaded.
Within the right end, an ORF of 366 aa had no homologue in the databases and a second ORF of 326 aa showed significant similarity to the fungal endonucleases S1 (32% identity, 45% similarity over 205 aa), P1, and PA3 (27–29). About 150 bp downstream of this ORF was a 505-bp region with 62% nucleotide identity to insertion sequences from Caulobacter crescentus (IS511) and Rhizobium meliloti (ISRm6) that are members of the IS3 family (30, 31). The translated sequence showed 60% identity to residues 99–263 of the 308-aa OrfB integrase of these IS sequences, but the element sequence was disrupted by a stop codon at the position corresponding to residue 203 and, in addition, there was no nucleotide or amino acid homology to the remainder of the IS OrfB. Hence it is unlikely that the element encodes an active integrase protein in this region. A further 183 bp downstream of the IS homology and 318 bp from the end of the element was a 35-bp sequence with 88% identity to the 3′ 35 bp of a methionine tRNA gene. This sequence was in the opposite orientation to the phe-tRNA located at the left junction (Fig. 3B). It seems possible that the partial met-tRNA gene and truncated IS integrase are relics that became part of the element as a result of previous integration or transfer events.
DISCUSSION
We have shown that the chromosomal symbiotic element of M. loti strain ICMP3153 is transmissible in laboratory matings to at least three genomic species of nonsymbiotic mesorhizobia. The 500-kb element integrates into a phe-tRNA gene and encodes an integrase of the phage P4 family just within its left end. The entire phe-tRNA gene is reconstructed at the left end of the element upon integration, whereas the 3′ 17 nucleotides of the tRNA gene are present as a direct repeat at the right end.
We propose the term symbiosis island for the element in analogy to the pathogenicity islands of Gram-negative bacterial pathogens. Like the symbiosis island, many pathogenicity islands are integrated adjacent to tRNA genes and convert environmental strains to strains able to form close associations with eukaryotic hosts (6–8). Other parallels between pathogenicity and symbiosis islands are that genes encoded on the islands are found in a broad range of species (32), and genes found on an island in one species may be located on a plasmid in another species. The symbiosis island differs from pathogenicity islands in that it is larger than any studied to date, comprising ≈10% of the genome of its host, and its transmission has been demonstrated.
Phage P4 and phiR73 integrate into tRNA genes with the 3′-terminal CCA defining the border of their attachment sites, such that tRNA function is not affected by integration or excision (33, 34). Integration of the symbiosis island within the phe-tRNA gene is likely to be mediated by the P4-related integrase in a similar fashion, with the 17-bp 3′ portion of the phe-tRNA comprising the island attachment site. The use of a highly conserved tRNA gene as the site of integration, and the transfer of the island to three genomic species, suggest that the island may have a broad host range. Two pathogenicity islands in E. coli strain J96, PAI-4 and PAI-5, are associated with phe-tRNA genes (35), whereas E. coli pathogenicity islands PAI-1, PAI-2, and LEE are associated with tRNA genes targeted by P4 (leuX) or phiR73 (selC) (32, 36). A conjugative transposon encoding sucrose utilization, CTnscr94, found in enteric bacteria also targets a phe-tRNA gene (37). We propose that the symbiosis island, CTnscr94, and pathogenicity islands represent a class of transmissible chromosomal genetic elements with features of phage, plasmids, and transposons. Further study of the symbiosis island should yield insight into its evolution and mode of transmission, and in more general terms, into mechanisms by which bacteria acquire clusters of genes such as pathogenicity islands that enable them to interact with eukaryotic hosts.
Acquisition of the symbiosis island converts a saprophyte into a symbiont. Therefore, as well as encoding genes required for nodule formation and symbiotic nitrogen fixation, the island is likely to contain other genes that contribute specifically to the success of the plant–microbe interaction. These genes may include genes required for the biosynthesis of biotin, nicotinate, and thiamin, as acquisition of the island converts the strains from auxotrophy to prototrophy for these vitamins. The absence of these functions in saprophytic nonsymbiotic mesorhizobia suggest they are not necessary and may be detrimental for a saprophytic lifestyle; their presence on the transferred DNA suggests they may contribute a competitive advantage in the presence of the host legume.
Acquisition of an island enables a bacterium to expand its genome to exploit new environmental niches. The reconstruction of the tRNA gene following integration provides the opportunity for acquisition of multiple islands in a tandem array and hence for the rapid diversification of bacterial populations. Such acquisition could account for the greater sequence variation found downstream of the insertion site among the strains examined compared with that found upstream (Fig. 2 and unpublished data), including between two strains R8CS and CJ4Sym that belong to the same species. Acquisition or loss of islands could also account for the divergence, apparently caused by gain or loss of large blocks of DNA, observed among environmental isolates of Pseudomonas aeruginosa (38). Certainly future studies of gene transfer between microbes in the environment should take account of the possibility that this transfer may be mediated by chromosomal islands.
Acknowledgments
This work was supported under Contract UOO601 from the New Zealand Foundation for Research, Science, and Technology, and by a grant from the Marsden Fund administered by the Royal Society of New Zealand.
Footnotes
References
- 1.Syvanen M. Annu Rev Genet. 1994;28:237–261. doi: 10.1146/annurev.ge.28.120194.001321. [DOI] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, et al. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Kolstø A-B. Mol Microbiol. 1997;24:241–248. doi: 10.1046/j.1365-2958.1997.3501715.x. [DOI] [PubMed] [Google Scholar]
- 4.Pace N R. Science. 1997;276:734–739. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 5.Covacci A, Falkow S, Berg D E, Rappuoli R. Trends Microbiol. 1997;5:205–208. doi: 10.1016/S0966-842X(97)01035-4. [DOI] [PubMed] [Google Scholar]
- 6.Lee C A. Infect Agents Dis. 1996;5:1–7. [PubMed] [Google Scholar]
- 7.Groisman E A, Ochman H. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 8.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 9.Groisman E A, Ochman H. Trends Microbiol. 1997;5:343–349. doi: 10.1016/S0966-842X(97)01099-8. [DOI] [PubMed] [Google Scholar]
- 10.Bäumler A J. Trends Microbiol. 1997;5:318–322. doi: 10.1016/S0966-842X(97)01082-2. [DOI] [PubMed] [Google Scholar]
- 11.Cheetham B F, Katz M. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 12.Cheetham B F, Tattersall D B, Bloomfield G A, Rood J I, Katz M E. Gene. 1995;162:53–58. doi: 10.1016/0378-1119(95)00315-w. [DOI] [PubMed] [Google Scholar]
- 13.Kovach M E, Shaffer M D, Peterson K M. Microbiology. 1996;142:2165–2174. doi: 10.1099/13500872-142-8-2165. [DOI] [PubMed] [Google Scholar]
- 14.Jarvis B D W, Van Berkum P, Chen W X, Nour S M, Fernandez M P, Cleyet-Marel J C, Gillis M. Int J Syst Bacteriol. 1997;47:895–898. [Google Scholar]
- 15.Pankhurst C E, MacDonald P E, Reeves J M. J Gen Microbiol. 1986;132:2321–2328. [Google Scholar]
- 16.Chua K-Y, Pankhurst C E, MacDonald P E, Hopcroft D H, Jarvis B D W, Scott D B. J Bacteriol. 1985;162:335–343. doi: 10.1128/jb.162.1.335-343.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan J T, Patrick H N, Lowther W L, Scott D B, Ronson C W. Proc Natl Acad Sci USA. 1995;92:8985–8989. doi: 10.1073/pnas.92.19.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan J T, Eardly B D, Van Berkum P, Ronson C W. Appl Environ Microbiol. 1996;62:2818–2825. doi: 10.1128/aem.62.8.2818-2825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ronson C W, Nixon B T, Albright L M, Ausubel F M. J Bacteriol. 1987;169:2424–2431. doi: 10.1128/jb.169.6.2424-2431.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y-N, Tang J-L, Clarke B R, Dow J M, Daniels M J. Mol Gen Genet. 1990;220:433–440. doi: 10.1007/BF00391750. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Clark C A, Beltrame J, Manning P A. Gene. 1991;107:43–52. doi: 10.1016/0378-1119(91)90295-m. [DOI] [PubMed] [Google Scholar]
- 24.Halling C, Calendar R, Christie G E, Dale E C, Deho G, et al. Nucleic Acids Res. 1990;18:1649. doi: 10.1093/nar/18.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirby J E, Trempy J E, Gottesman S. J Bacteriol. 1994;176:2068–2081. doi: 10.1128/jb.176.7.2068-2081.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun J, Inouye M, Inouye S. J Bacteriol. 1991;173:4171–4181. doi: 10.1128/jb.173.13.4171-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maekewa K, Tsunasawa S, Dibo G, Sakiyama F. Eur J Biochem. 1991;200:651–661. doi: 10.1111/j.1432-1033.1991.tb16228.x. [DOI] [PubMed] [Google Scholar]
- 28.Lee B R, Kitamoto K, Yamada O, Kumagai C. Appl Microbiol Biotechnol. 1995;44:425–431. doi: 10.1007/BF00169939. [DOI] [PubMed] [Google Scholar]
- 29.Tabata N, Kazama H, Ohgi K, Irie M. Agric Biol Chem. 1991;55:461–469. [PubMed] [Google Scholar]
- 30.Mullin D A, Zies D L, Mullin A H, Caballera N, Ely B. Mol Gen Genet. 1997;254:456–463. doi: 10.1007/s004380050439. [DOI] [PubMed] [Google Scholar]
- 31.Zekri S, Toro N. Gene. 1996;175:43–48. doi: 10.1016/0378-1119(96)00118-7. [DOI] [PubMed] [Google Scholar]
- 32.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inouye S, Sunshine M G, Six E W, Inouye M. Science. 1991;252:969–971. doi: 10.1126/science.1709758. [DOI] [PubMed] [Google Scholar]
- 34.Pierson L S, Kahn M L. J Mol Biol. 1987;196:487–496. doi: 10.1016/0022-2836(87)90026-x. [DOI] [PubMed] [Google Scholar]
- 35.Swenson D L, Bukanov N O, Berg D E, Welch R A. Infect Immun. 1996;64:3736–3743. doi: 10.1128/iai.64.9.3736-3743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschäpe H, Hacker J. Infect Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hochhut B, Jahreis K, Lengeler J W, Schmid K. J Bacteriol. 1997;179:2097–2102. doi: 10.1128/jb.179.7.2097-2102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Römling U, Schmidt K D, Tümmler B. J Mol Biol. 1997;271:386–404. doi: 10.1006/jmbi.1997.1186. [DOI] [PubMed] [Google Scholar]