Abstract
We have identified and characterized ARC, apoptosis repressor with caspase recruitment domain (CARD). Sequence analysis revealed that ARC contains an N-terminal CARD fused to a C-terminal region rich in proline/glutamic acid residues. The CARD domain of ARC exhibited significant homology to the prodomains of apical caspases and the CARDs present in the cell death regulators Apaf-1 and RAIDD. Immunoprecipitation analysis revealed that ARC interacts with caspase-2, -8, and Caenorhabditis elegans CED-3, but not with caspase-1, -3, or -9. ARC inhibited apoptosis induced by caspase-8 and CED-3 but not that mediated by caspase-9. Further analysis showed that the enzymatic activity of caspase-8 was inhibited by ARC in 293T cells. Consistent with the inhibition of caspase-8, ARC attenuated apoptosis induced by FADD and TRADD and that triggered by stimulation of death receptors coupled to caspase-8, including CD95/Fas, tumor necrosis factor-R1, and TRAMP/DR3. Remarkably, the expression of human ARC was primarily restricted to skeletal muscle and cardiac tissue. Thus, ARC represents an inhibitor of apoptosis expressed in muscle that appears to selectively target caspases. Delivery of ARC by gene transfer or enhancement of its endogenous activity may provide a strategy for the treatment of diseases that are characterized by inappropriately increased cell death in muscle tissue.
Keywords: cell death
Apoptosis, a morphologically distinguished form of programmed cell death, is critical not only during development and tissue homeostasis but also in the pathogenesis of a variety of diseases including cancer, autoimmune disease, viral infection, and degenerative disorders (1, 2). Several regulatory components of the apoptotic pathway have been identified in various living organisms including humans (3, 4). In mammals, a family of cysteine proteases (designated caspases) related to the Caenorhabditis elegans CED-3 protein appears to represent a major effector arm of the apoptotic program (5). To date, more than 10 caspases have been identified and partially characterized (6). Several of these caspases, notably caspase-2, -3, -4, -6, -7, -8, -9, and -10 have been implicated in the induction of apoptosis (6). The caspases are synthesized as inactive precursors that are proteolytically processed to generate active subunits. Each caspase contains conserved sequences important for proteolytic activity cleaving after specific aspartic acid residues (6). The mammalian cell death proteases have been divided into proximal and distal caspases based on the their sites of action in the proteolytic caspase cascade (6). Activation of apical caspases, such as caspase-8, through cell death receptors or other apoptotic stimuli leads to activation of downstream caspases, precipitous cleavage of target proteins and execution of the apoptotic program (7, 8).
Little is known about the regulation of caspase activity during apoptosis. In the nematode C. elegans, activation of the cell death protease CED-3 is positively regulated by CED-4 and inhibited by CED-9 through direct protein-protein interactions (9, 10). Likewise, Apaf-1, a human protein that resembles C. elegans CED-4, interacts with caspase-9, a step that is required for the activation of the downstream protease caspase-3 (11). The prodomains of several apical caspases contain a protein module termed caspase recruitment domain (CARD) that is conserved in several apoptosis regulatory molecules, including Apaf-1, RAIDD, and cellular inhibitors of apoptosis proteins (IAPs) (12). The CARD has been proposed to play a regulatory role in apoptosis by allowing proteins such as Apaf-1 to associate with caspase-9 (13). Two viral proteins, baculovirus p35 and cowpox virus CrmA, inhibit apoptosis by directly targeting caspases (14, 15). The IAPs comprise a family of apoptosis inhibitors found in baculoviruses, Drosophila, and mammals (16, 17). Mammalian IAP-1, -2, and XIAP directly bind and inhibit enzymatically active death proteases, caspase-3, and -7, but not the upstream protease caspase-8 (18, 19).
Apoptosis has been proposed to play a role in the development and/or progression of several inherited and acquired diseases of the skeletal and cardiac muscle (20–23). However, little is known about the molecular regulation of apoptosis in muscle cells. In the current study, we have identified and characterized a human cDNA encoding an apoptosis repressor with a CARD (ARC) that is expressed in skeletal muscle and heart. ARC interacts selectively with caspases and functions as an inhibitor of apoptosis.
MATERIALS AND METHODS
Isolation of ARC and Construction of Expression Plasmids.
The partial nucleotide sequences of cDNAs encoding peptides with homology to the CARD of caspase-9 (amino acids 1–80) were found in expressed-sequence tag (EST) databases of GenBank using the tblastn program. The entire nucleotide sequence of a cDNA containing a 1.0-kb insert corresponding to EST clones 322821, 546171 and 588443 was determined by dideoxy-sequencing. The entire ORF of ARC from EST clone 322821 was tagged at the C terminus with Flag or hemagglutinin (HA) sequences and cloned into the expression vector pcDNA3 (Invitrogen) to produce pcDNA3-ARC-Flag or pcDNA3-ARC-HA. The human caspase-8 (amino acids 1–215) and caspase-8 (amino acids 216–496) were fused at the C terminus with HA-tag sequences and cloned into pcDNA3 to produce pcDNA3-N-caspase-8-HA or pcDNA3-C-caspase-8-HA, respectively. The authenticity of all constructs was confirmed by dideoxy sequencing. pcDNA3-caspase-1-Flag, pcDNA3-caspase-3-Flag, pcDNA3-caspase-8-AU1, pcDNA3-caspase-8-mt(C377S)-AU1, pcDNA3-caspase-9-Flag, pcDNA3-Ced-3-Flag, pcDNA3-FADD-AU1, pcDNA3-HA-TRADD pcDNA3-CLARP-myc, and pcDNA3-p35 were described (10, 24–25). pcDNA3-FAS and pcDNA3-TNFR1-Flag were gifts from V. Dixit (Genentech). pcDNA3-TRAMP was a gift from J. Tschopp (University of Lausanne).
Northern Blot Analysis.
A 1.0-kb fragment containing the ARC coding sequence was radiolabeled by random priming using a commercial kit (Boehringer Mannheim) and applied for analysis of human multiple poly(A)+ mRNA blots (CLONTECH) according to the manufacturer’s instructions.
Transfection, Expression, Immunoprecipitation, and Immunodetection of Tagged Proteins.
5 × 106 human 293T cells were transfected with expression plasmids by a calcium phosphate method as described (24). Briefly, 2 μg of pcDNA3 or pcDNA3-ARC-Flag or -HA was cotransfected with 3 μg of pcDNA3 or caspase expression plasmids. Total amount of transfected plasmid DNA was always 5 μg. 293T cells were harvested after 22 hr and lysed with 0.2% Nonidet P-40 isotonic lysis buffer (26). For immunoprecipitation, 1 μg of soluble protein was incubated with 10 μg/ml of polyclonal anti-Flag, monoclonal anti-AU1, or polyclonal anti-HA antibody for 2 hr at 4°C and tagged proteins were immunoprecipitated with protein A-Sepharose 4B (Zymed). Immunoprecipitates were subjected to SDS/12% polyacrylamide gel and immunoblotted with anti-Flag, anti-AU1, or anti-HA antibodies.
β-Galactosidase Apoptosis Assay and Caspase-8 Enzymatic Assay.
5 × 105 293T cells were transfected with 0.3 μg of pcDNA3-β-galactosidase plus each expression plasmid in triplicate. Cells were fixed 16 hr after transfection, stained for β-galactosidase and assayed for morphological features of apoptosis (27). Statistical significance was determined by one-way ANOVA followed by Student-Neuman-Keuls post hoc comparisons. For caspase-8 enzymatic assay, 1.5 × 106 human 293T cells were cotransfected with pcDNA3, pcDNA3-ARC-Flag, pcDNA3-caspase-8, and pcDNA3-caspase-8-mt by a calcium phosphate method. Total amount of transfected plasmid DNA was always 2.5 μg. 293T cells were harvested 18 hr after transfection and lysed with 0.2% Nonidet P-40 isotonic lysis buffer. Tagged proteins were immunoprecipitated and caspase activity was measured as described (25).
RESULTS
Identification of ARC, a Human Protein with Homology to the Prodomains of Caspases and Apaf-1.
To identify apoptosis-regulatory proteins, we screened the GenBank database for cDNAs encoding proteins with homology to the CARD of caspase-9 (amino acid residues 1–80) by computer homology search. Several human ESTs containing overlapping nucleotide sequences with significant amino acid homology to caspase-9 were identified. The longest cDNA (EST clone 322821) was 1.0 kb, and its nucleotide sequence revealed an ORF that encoded a protein of 208 amino acids with a predicted relative molecular mass of 22,629 Da (Fig. 1). We designated this human protein as ARC (apoptosis repressor with CARD, see below). The amino acid sequence of human ARC was highly homologous (82% identical) to a predicted 221 amino acid rat protein of unknown function whose cDNA was identified through a screening for proteins containing glutamate-proline repeats (28). Thus, the rat clone appears to represent the rat ortholog of human ARC. Alignment analysis revealed that both human and rat ARC are proteins containing an N-terminal CARD (12) and a C-terminal region rich in proline and glutamic acid residues (Fig. 1B). The CARD of human and rat ARC have significant amino acid similarity to the CARDs from caspase-2, caspase-9, RAIDD, and Apaf-1 (Fig. 1C).
Figure 1.
Structure, sequence, and alignment of ARC with related proteins. (A) Schematic structure of human ARC. CARD and proline/glutamic acid-rich domains are shown as closed and open boxes. (B) The amino acid sequences of human and rat ARC are aligned. The identical residues in human and rat ARC were indicated by asterisk. (C) Alignment of the amino acid sequences of CARD domains of ARC, caspase-9, human caspase-2, RAIDD, and Apaf-1. The conserved residues in human and rat ARC were indicated by asterisk.
Human ARC Is Expressed in Skeletal Muscle and Heart.
We performed Northern blot analysis to assess the expression of arc mRNA in various human tissues. Hybridization with an ARC probe showed two transcripts of ≈5.5 kb and 1.0 kb in skeletal muscle and heart but not in brain, placenta, lung, liver, kidney, pancreas, or various lymphoid-hematopoietic tissues (Fig. 2). The 1.0-kb transcript represents the cDNA analyzed in the present study. The significance and identity of the 5.5-kb mRNA transcript is unclear. It could represent a RNA form of ARC derived by alternative splicing, usage of an alternative polyadenylation sites, or cross-hybridization of the probe with sequences of a related gene.
Figure 2.
Expression of ARC in human tissues by Northern blot analysis. Poly(A)+ RNAs from various tissues were hybridized with a probe corresponding to the entire human ARC cDNA.
Overexpression of ARC Inhibits Apoptosis Induced by Caspases in 293T Cells.
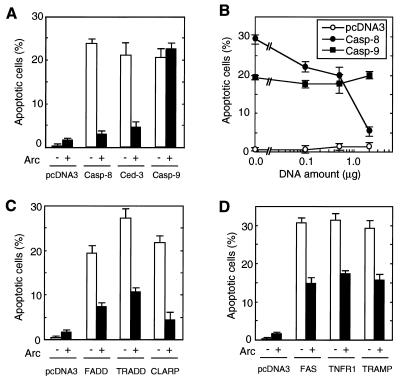
To begin to elucidate the physiological function of ARC, an expression construct producing ARC was introduced into human kidney epithelial 293T cells and subsequently observed for features of apoptosis. Expression of ARC did not induce apoptosis of 293T cells (data not shown). Because the N-terminal region of ARC exhibited homology to the prodomains of several apical caspases, we reasoned that ARC might regulate the killing activity of caspases. To test that, plasmids producing several caspases known to activate cell death were coexpressed with ARC in 293T cells. Expression of ARC inhibited apoptosis induced by caspase-8 and C. elegans CED-3 (P < 0.01) but not that mediated by caspase-9 (Fig. 3A). Further experiments revealed that ARC inhibited caspase-8-induced killing in a dose-dependent manner (Fig. 3B).
Figure 3.
ARC is a negative regulator of apoptosis. 293T cells were transfected with pcDNA3, pcDNA3-ARC-Flag, and various expression plasmids as described in Material and Methods. Transfected cells were visualized with β-galactosidase substrate and scored for morphological feature of apoptosis. (A) Caspases were cotransfected with ARC (▪) or without ARC (□). In the experiment, 0.2 μg of plasmid DNA expressing caspase-4, -8, or -10 or 0.1 μg of Ced-3 or caspase-9 plasmid was used in the presence of 2 μg of pcDNA3-ARC-Flag or pcDNA3. (B) ARC inhibits caspase-induced apoptosis in a dose-dependent manner. 0.2 μg of plasmid expressing caspase-4, -8, -10, or pcDNA3 was used. The x axis represents amount of ARC plasmid DNA. Total amounts of plasmid DNA was 2.2 μg in all experiments presented in A and B. (C) ARC inhibits FADD, TRADD, and CLARP-induced apoptosis. The amount of plasmid DNA were: FADD (0.4 μg), TRADD (0.1 μg), CLARP (2.0 μg), or pcDNA (2.0 μg) in the presence of 2.0 μg of ARC or pcDNA3 plasmid. (D) ARC inhibits apoptosis induced by death receptors. The amount of plasmid DNA were: Fas (1.5 μg), TNFR1 (0.2 μg), TRAMP (1.0 μg), or pcDNA3 (2.0 μg) in the presence of 2.0 μg of ARC or pcDNA3 plasmid. Total amounts of plasmid DNA was 4.0 μg in all experiments presented in C and D.
ARC Inhibits Apoptosis Mediated by Stimulation of Death Receptor Pathways.
Stimulation of several members of the tumor necrosis factor (TNF) family of receptors including TNF-R1, CD95/Fas, and TRAMP/DR3 induces apoptosis through engagement of the apical protease caspase-8 (7–8, 29). We performed experiments to assess the regulation by ARC of apoptosis induced by signaling molecules that function upstream of caspase-8 in the death receptor pathways. Fig. 3C shows that ARC inhibited apoptosis induced by FADD and TRADD, two signaling molecules of CD95/Fas and TNF-R1 pathways respectively (P < 0.01), whose stimulation leads to activation of caspase-8 and apoptosis (30–33). In addition, ARC inhibited apoptosis induced by CLARP, a caspase-like protein that interacts with caspase-8 (25). Consistent with the results shown in Fig. 3C, expression of ARC partially but significantly inhibited apoptosis induced by stimulation of CD95/Fas, TNF-R1, and TRAMP/DR3 receptors (P < 0.01) (Fig. 3D).
ARC Inhibits the Enzymatic Activity of Caspase-8.
The experiments shown above indicate that ARC inhibits apoptosis induced by several caspases, including caspase-8. We performed experiments to test whether ARC could regulate the enzymatic activity of caspase-8, a function that is required for caspase-8 to activate apoptosis (7, 8). To examine if ARC regulates the enzymatic activity of caspase-8 in intact cells, 293T cells were transiently transfected with expression plasmids producing Flag-tagged caspase-8 and ARC or control plasmid. Caspase-8 was immunoprecipitated with anti-Flag antibody and the immunoprecipitates were assayed for caspase activity by using the fluorogenic substrate acetyl-Asp-Glu-Val-Asp7-amino-4-methylcoumarin. Enzymatic analysis showed that ARC inhibited the enzymatic activity of caspase-8 (Fig. 4A). In control experiments, immunoprecipitates from cells transfected with control plasmid or constructs expressing ARC alone or a caspase-8 mutant with a single amino acid change (cysteine to serine) in the conserved active pentapeptide did not exhibit detectable enzymatic activity (Fig. 4A). Immunoblotting with anti-Flag antibody revealed that extracts assayed for caspase activity expressed similar levels of caspase-8 (Fig. 4B).
Figure 4.
ARC suppresses the enzymatic activity of caspase-8 in intact cells. (A) 293T cells were cotransfected with 0.2 μg of pcDNA3-caspase-8-AU1 or pcDNA3-caspase-8-mut and 2 μg of pcDNA3-ARC-Flag or pcDNA3. Caspase-8 in cell extracts was immunoprecipitated with anti-AU1 antibody and immunoprecipitates were incubated with the fluorogenic substrate acetyl-Asp-Glu-Val-Asp7-amino-4-methylcoumarin. ○, pcDNA3 alone; •, pcDNA3-ARC-HA alone; □, pcDNA3-caspase-8-AU1 alone; ▪, pcDNA3-caspase-8-AU1 and pcDNA3-ARC-HA; ▵, pcDNA3-caspase-8-mt-AU1 alone. (B) AU1-tagged caspase-8 and caspase-8-mt were detected in immunoprecipitates with anti-AU1 by immunoblotting.
ARC Interacts with Caspase-2, Caspase-8, and C. elegans CED-3 but Not with Caspase-1, -3, or -9.
The inhibition of caspase-mediated function by ARC suggested that ARC might physically interact with caspases. To determine if ARC associates with caspases, we transiently cotransfected 293T cells with expression plasmids producing caspase-1, caspase-2, caspase-3, caspase-8, caspase-9, C. elegans CED-3, or control empty vector and Flag or HA tagged ARC. Immunoblotting of ARC complexes immunoprecipitated with anti-Flag antibody revealed that ARC was coimmunoprecipitated with caspase-2, -8, and CED-3 but not with caspase-1, -3, or -9 (Fig. 5 A–C). Analysis of total lysates by immunoblotting revealed that lack of interaction between ARC and caspase-1, -3, or -9 was not due to inappropriate expression of these proteins in cell extracts (Fig. 5 B and C). Further analysis of caspase-8 deletion mutants revealed that ARC associated with the N-terminal death effector domain but not with the C-terminal region that contains the catalytic domain of caspase-8 (Fig. 5D). Furthermore, ARC did not associate with several apoptosis regulatory molecules including FADD, RAIDD, Bcl-XL, and c-IAP-2 (data not shown), further supporting the specificity of the ARC interactions.
Figure 5.
ARC interacts with caspase-2, -8, and Ced-3 but not with caspase-1, -3, and -9. (A) 293T cells were transfected with plasmids AU1-tagged caspase-2 or -8 and HA-tagged ARC. Lysates were immunoprecipitated with anti-AU1 antibody and immunoblotted with anti-HA antibody (Top). Total lysates (100 μg) were immunoblotted with anti-AU1 (Center) or anti-HA antibody (Bottom). (B and C) 293T cells were transfected with plasmids Flag-tagged caspase-1, -3, -9, or CED-3 and HA-tagged ARC. Lysates were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-HA antibody (Top). Total lysates (100 μg) were immunoblotted with anti-Flag (Center) or anti-HA antibody (Bottom). (D) 293T cells were transfected with plasmids HA-tagged N-caspase-8 or C-caspase-8, and Flag-tagged ARC. Lysates were immunoprecipitated with anti-HA antibody and immunoblotted with anti-Flag antibody (Top). Total lysates (100 μg) were immunoblotted with anti-HA (Center) or anti-Flag antibody (Bottom).
DISCUSSION
In the current work we describe ARC, a protein containing a CARD that functions as an inhibitor of apoptosis. The inhibitory effect of ARC is selective in that it repressed apoptosis induced by caspase-8 and C. elegans CED-3, but not that mediated by caspase-9. ARC inhibited apoptosis mediated by stimulation of death receptors such as CD95/Fas, TNFR1, and TRAMP, as well as that activated by FADD and TRADD, two signaling molecules of the CD95/Fas and TNFR1 pathways whose expression can activate apoptosis (30–34). Because these death receptors as well as FADD and TRADD mediate their apoptotic effect through the activation of the apical proteases caspase-8 and/or caspase-2 (30–33), ARC is likely to regulate death receptor-induced apoptosis via its interactions with caspase-2 and caspase-8.
The mechanism by which ARC inhibits apoptosis remains unclear and needs to be further investigated. There are at least two possible models that could explain the apoptosis inhibitory function of ARC. (i) ARC might repress apoptosis by inhibiting caspase activation through direct binding to death proteases. ARC could act by inhibiting the processing of immature caspases and/or direct inhibition of the active caspase subunits. The observation that ARC did not interact with the C-terminal region that contains the catalytic domains of caspase-8 suggests that ARC acts by targeting the immature caspase form. Cleavage of ARC was not observed when interacting with caspases, implying a mechanism different from that of the baculovirus p35 protein (14). Three mammalian IAPs—XIAP, c-IAP-1, and c-IAP-2—have been shown to interact with and inhibit specific caspases (18, 19). Unlike ARC, however, cellular IAPs target distal death proteases such as caspase-3 and caspase-7, but not the proximal protease caspase-8 (18, 19). (ii) ARC could inhibit apoptosis by disrupting the association between death proteases and their activators such as FADD or RAIDD. A similar mechanism has been proposed for FLIP proteins, a caspase-related molecule that like ARC interacts with caspase-8 (35). The interaction between ARC and caspases appear to be mediated via the corresponding CARD or the structurally related death effector domains. Thus, ARC associated with caspase-2, -8, and CED-3, but not with caspase-3, a death protease that lacks such a domain (6). Furthermore, mutant analysis of caspase-8 showed that the N-terminal region containing death effector domains was required for its interaction with ARC. However, ARC did not interact with CARD-containing caspase-1 or -9. Significantly, RAIDD has been reported to bind caspase-2 but not caspase-1 (36) although both caspases have CARD domains. These observations suggest that subtle differences may exist among individual CARD domains or that other factors play a critical role in these interactions.
The expression of ARC was highly restricted to skeletal muscle and heart suggesting that ARC plays a role in the regulation of apoptosis in muscle tissues. Striated myofibers in skeletal muscle and heart are long-lived cells. However, little is known about the mechanisms that inhibit apoptosis in muscle cells and are responsible for their long-term survival. Bcl-2 and Bcl-XL, two members of the Bcl-2 family, promote survival but they are expressed at low or undetectable levels in skeletal muscle (37). Thus, ARC expression may play a role in maintaining myofiber survival in skeletal muscle and heart tissues. However, a role for ARC in muscle cell survival remains to be investigated by analysis of ARC in muscle cells. Several inherited diseases including muscular dystrophy and spinal muscle atrophy are characterized by degeneration of muscle fibers through apoptosis and necrosis (20–21). Furthermore, dystrophic muscle of the mdx mouse and BIO14.6 hamster undergo apoptosis, degeneration, and subsequently necrosis as disease progresses (37–38). Similarly, acquired conditions such as inflammatory myopathies, myocardial infarction and overload-induced myopathy have been shown to have a component of apoptotic cell death (22, 23, 39, 40). It is conceivable therefore that ARC could regulate apoptosis associated with these muscle cell diseases. In addition, expression of ARC could provide a novel therapeutic approach that could be accomplished by direct delivery of ARC to the areas of insult via gene therapy or through drugs capable of enhancing the activity or expression of endogenous ARC.
Acknowledgments
We thank M. Benedict, Y. Hu, D. Wu, M. Gonzalez-Garcia, and L. del Peso for critical review of the manuscript and E. Alnemri, V. Dixit, and J. Tschopp for plasmids. This work was supported by National Institutes of Health Grant R01 CA64556–01. T.K. was supported by a fellowship from the Japan Science and Technology Corporation. G.N. was supported by a Research Career Development Award K04 CA64421–01 from the National Institutes of Health.
ABBREVIATIONS
- CARD
caspase recruitment domain
- ARC
apoptosis repressor with CARD
- IAP
inhibitors of apoptosis protein
- TNF
tumor necrosis factor
- EST
expressed-sequence tags
- HA
hemagglutinin
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. AF043244).
References
- 1.Vaux D L, Strasser A. Proc Natl Acad Sci USA. 1996;93:2239–2244. doi: 10.1073/pnas.93.6.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson C B. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 3.White E. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Jacobson M D, Weil M, Raff M C. Cell. 1997;88:347–54. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 5.Alnemri E S, Livingston D J, Nicholson D W, Salvesen G, Thornberry N A, Wong W W, Yuan J. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 6.Cohen G M. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 8.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, et al. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 9.Chinnaiyan A M, O’Rourke K, Lane B R, Dixit V M. Science. 1997;275:1122–1126. doi: 10.1126/science.275.5303.1122. [DOI] [PubMed] [Google Scholar]
- 10.Wu D, Wallen H D, Inohara N, Núñez G. J Biol Chem. 1997;272:21449–21454. doi: 10.1074/jbc.272.34.21449. [DOI] [PubMed] [Google Scholar]
- 11.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann K, Bucher P, Tschopp J. Trends Biochem Sci. 1997;22:155–156. doi: 10.1016/s0968-0004(97)01043-8. [DOI] [PubMed] [Google Scholar]
- 13.Li P., Nijhawan, D., Budihardjo, I., Srinivasula, S. M., Ahmad, M., Alnemri, E. S. & Wang, X. (1997) Cell 91 (479–489). [DOI] [PubMed]
- 14.Bump N J, Hackett M, Hugunin M, Seshagriri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Licari L P, et al. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Q, Snipas S, Orth K, Muzio M, Dixit V M, Salvesen G S. J Biol Chem. 1997;272:7797–7800. doi: 10.1074/jbc.272.12.7797. [DOI] [PubMed] [Google Scholar]
- 16.Crook N E, Clem R J, Miller L K. J Virol. 1993;67:2168–74. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duckett C S, Nava V E, Gedrich R W, Clem R J, Van Dongen J L, Gilfllan M C, Shiels H, Hardwick J M, Thompsom C B. EMBO J. 1996;15:2695–2694. [PMC free article] [PubMed] [Google Scholar]
- 18.Devereaux Q L, Takahashi R, Salvesen G S, Reed J C. Nature (London) 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 19.Roy N, Deveraux Q L, Takahashi R, Salvesen G S, Reed J C. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tews D S, Goebel H H. J Neuropathol Exp Neurol. 1997;56:150–156. doi: 10.1097/00005072-199702000-00005. [DOI] [PubMed] [Google Scholar]
- 21.Fidzianska A, Goebel H H, Warlo I. Brain. 1990;113:433–445. doi: 10.1093/brain/113.2.433. [DOI] [PubMed] [Google Scholar]
- 22.Bialik S, Geene D L, Sasson I E, Cheng R, Horner J W, Evams S M, Lord E M, Knok C J, Kitsis R N. J Clin Invest. 1997;100:1363–1372. doi: 10.1172/JCI119656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olivetti G, Abbi R, Quaini F, Kajstura J, Chang W, Nitahara J A, Quaini E, DiLoreto C, Beltrami C A, Krajewski S, et al. N Engl J Med. 1997;336:1131–1141. doi: 10.1056/NEJM199704173361603. [DOI] [PubMed] [Google Scholar]
- 24.Inohara N, Ding L, Chen S, Núñez G. EMBO J. 1997;16:1686–1694. doi: 10.1093/emboj/16.7.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inohara N, Koseki T, Hu Y, Chen S, Núñez G. Proc Natl Acad Sci USA. 1997;94:10717–10722. doi: 10.1073/pnas.94.20.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oltvai Z N, Milliman C L, Korsmeyer S J. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 27.Miura M, Zhu H, Rotello R, Hartwieg E A, Yuan J. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 28.Geertman R, McMahan A, Sabban E L. Biochim Biophys Acta. 1996;1306:147–152. doi: 10.1016/0167-4781(96)00036-x. [DOI] [PubMed] [Google Scholar]
- 29.Chinnaiyan A M, O’Rourke K, Yu G-L, Lyons R H, Garg M, Duan D R, Xing L, Gentz R, Ni J, Dixit V M. Science. 1996;274:990–992. doi: 10.1126/science.274.5289.990. [DOI] [PubMed] [Google Scholar]
- 30.Chinnaiyan A M, O’Rourke K, Tewari M, Dixit V M. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 31.Hsu H, Xiong J, Goeddel D V. Cell. 1995;19:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 32.Kischkel F C, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer P H, Peter M E. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chinnaiyan A M, Tepper C G, Seldin M F, O’Rourke K, Kischkel F C, Hellbardt S, Krammer P H, Peter M E, Dixit V M. J Biol Chem. 1996;271:4961–4965. doi: 10.1074/jbc.271.9.4961. [DOI] [PubMed] [Google Scholar]
- 34.Itoh N, Yonehara S, Ishii A, Yonehara M, Mizushima S, Sameshima M, Hase A, Seto Y, Nagata S. Cell. 1991;26:233–243. doi: 10.1016/0092-8674(91)90614-5. [DOI] [PubMed] [Google Scholar]
- 35.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer J L, Schroter M, Burns K, Mattmann C, et al. Nature (London) 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 36.Duan H, Dixit V M. Nature (London) 1997;385:86–89. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- 37.Matsuda R, Nishikawa A, Tanaka H. J Biochem. 1995;118:959–964. doi: 10.1093/jb/118.5.959. [DOI] [PubMed] [Google Scholar]
- 38.Tidbell J G, Albrecht D E, Lokensgard B E, Spencer M J. J Cell Sci. 1995;108:2197–2204. doi: 10.1242/jcs.108.6.2197. [DOI] [PubMed] [Google Scholar]
- 39.Behrens L, Bander A, Johnson M A, Hohlfeld R. Brain. 1997;120:929–938. doi: 10.1093/brain/120.6.929. [DOI] [PubMed] [Google Scholar]
- 40.Teiger E, Than V D, Richard L, Wisnewsky C, Tea B S, Gaboury L, Tremblay J, Schwartz K, Hamet P. J Clin Invest. 1996;97:2891–2897. doi: 10.1172/JCI118747. [DOI] [PMC free article] [PubMed] [Google Scholar]