Abstract
The spontaneous mouse mutant Dominant megacolon (Dom) is a valuable model for the study of human congenital megacolon (Hirschsprung disease). Here we report that the defect in the Dom mouse is caused by mutation of the gene encoding the Sry-related transcription factor Sox10. This assignment is based on (i) colocalization of the Sox10 gene with the Dom mutation on chromosome 15; (ii) altered Sox10 expression in the gut and in neural-crest derived structures of cranial ganglia of Dom mice; (iii) presence of a frameshift in the Sox10 coding region, and (iv) functional inactivation of the resulting truncated protein. These results identify the transcriptional regulator Sox10 as an essential factor in mouse neural crest development and as a further candidate gene for human Hirschsprung disease, especially in cases where it is associated with features of Waardenburg syndrome.
Keywords: neural crest, high-mobility-group domain, transcription, glia
Cells of the neural crest give rise to a plethora of functionally diverse cell types, including neurons, glia, neuroendocrine cells, and melanocytes (1). Considerable insight into the cellular and molecular mechanisms of neural crest development has come from studies of spontaneous mouse mutants such as lethal spotting (ls), piebald-lethal (sl) and Dominant megacolon (Dom) (2, 3). Similar to the autosomal recessive ls and sl mutations, the semidominant Dom mutation affects several aspects of neural crest development, leading to combined intestinal aganglionosis and pigmentation defects. Both enteric hypo- or aganglionosis and spotted pigmentation vary in Dom/+ mice with the genetic background. The majority of Dom/Dom homozygous embryos die before embryonic day 13 (E13) (3). However, on a C57BL/6J × C3H/HheOuJ hybrid genetic background, very few homozygous embryos develop to term and newborn Dom/Dom pups die within a few hours after birth.
Whereas the underlying genetic defects in the autosomal recessive ls and sl mutations are well characterized and have been shown to result from functional inactivation of the endothelin-3 and endothelin receptor B gene, respectively (4, 5), the molecular basis of the semidominant Dom phenotype is unknown. Here we identify the defect in Dom mice to be caused by mutation of Sox10, a recently identified member of the Sox gene family of transcriptional regulators with prominent expression in the early neural crest, the developing enteric nervous system (ENS), and in glial cells of the peripheral and central nervous systems (6).
All members of the Sox gene family are characterized by possession of a DNA-binding domain with similarity to the high-mobility group (HMG)-domain of the mammalian sex-determining gene SRY (7). They have a strong DNA-bending capacity and are thought to function as architectural proteins that shape the three-dimensional conformation of multiprotein-DNA complexes on promoters and enhancers (8). Sox genes exist in such evolutionary distant species as human and nematode and are involved in developmental processes as diverse as sex determination, hemopoiesis, chondrogenesis (7), and (as shown here for Sox10) neural crest development.
EXPERIMENTAL PROCEDURES
Sox10 Mapping, Sequencing, and Expression.
The genetic map of the Dom region was established by using a total of 528 Dom/+ mice. All used gene polymorphisms between Mus spretus and C57BL/6J are described (9) except for H1Fv and Sox10. Primers 1 (5′-ATGACTCCTTTTTCCTTAAC-3′) and 2 (5′-CAAGCAGTCTAAGAAAGTCT-3′) generate a single strand conformational polymorphism for H1Fv. Primers 3 (5′-CAGCTGGTTCTGTCCTGTCA-3′) and 4 (5′-GAAGAAGGCTGGGTGGATTG-3′) produce a fragment length polymorphism for Sox10. Yeast artificial chromosomes (YACs) were identified from the St Mary’s Hospital library, London (B24D4), the Imperial Cancer Research Fund library, London (C3H2D4 and C3H15D5), and the Princeton mouse YACs library (142G11 and 193F1) (10–12). Right ends and left ends were isolated by bubble PCR and used to generate new markers. YAC chimerism was tested by using the mouse/hamster hybrid panel provided by the United Kingdom Human Genome Mapping Project Resource Center (13).
The nucleotide sequence for the entire coding region of mouse Sox10 and its Dom variant were determined in a two-step reverse transcription–PCR on total brain RNA from C57BL/6J and homozygous Dom newborn and were submitted to EMBL/GenBank (accession nos. AF047043 and AF047389). Both the glutamate11-to-valine substitution and the detected frameshift were separately introduced into the rat Sox10 cDNA by site-directed mutagenesis. The altered Sox10 cDNAs subsequently were inserted in the eukaryotic expression plasmid pCMV5 in a manner similar to wild-type Sox10 (6) and expressed in COS cells after transient transfection by the DEAE dextran technique using a concentration of 500 μg/ml DEAE dextran followed by chloroquine treatment. Nuclear extracts were prepared 2 days posttransfection and used in Western blotting experiments with a Sox10-specific rabbit antiserum (6).
Genotyping, RNA Preparation, and Northern Blotting.
Mice were genotyped by using the D15Mit71 microsatellite, which generates a 14-bp polymorphism between C57BL/6J and C3H/HeOuJ. Total RNA from dissected mouse tissues was isolated by using Trizol reagent (GIBCO/BRL), separated on formaldehyde-containing 1% agarose gels, and blotted onto Duralose-UV membranes (Stratagene). Filters were hybridized to 32P random-labeled probes for Sox10 and β-actin as described (6).
Immunohistochemistry and in Situ Hybridization.
E10.5 embryos were subjected to whole-mount immunohistochemistry by using the anti-NF160 mouse monoclonal 2H3 (Jessell and Dodd, Developmental Studies Hybridoma Bank, Iowa City) as described (14). In situ hybridization of whole mounts or 10-μm cryostat sections was performed by using DIG-UTP- and 35S-UTP labeled riboprobes, respectively (6). The second half of the rat Sox10 cDNA (positions 1287–3030 according to GenBank accession no. AJ001029) without the HMG-domain was transcribed into the 1.75-kb antisense riboprobe. No signal was obtained for the corresponding sense probe.
Functional Assays.
Electrophoretic mobility-shift assays were performed with 0.4 μg of nuclear extract from transiently transfected COS cells and 0.5 ng of 32P-labeled consensus Sox binding site 5′-GATCCGCGCCTTTGTTCTCCCCA-3′ (6). Transcriptional activities were determined in U138 glioblastoma cells transiently transfected by the calcium phosphate technique using Sox10-containing and other previously described expression plasmids (0.2 μg per 60-mm plate) and luciferase reporter plasmids (2 μg per 60-mm plate) (6). The 3xFXO luc and 3xFXP luc reporters carry three tandem copies of the FXO sequence (consisting of adjacent binding sites for Sox and POU proteins) or FXP sequence (consisting of adjacent binding sites for Sox and Pax proteins) in front of the β-globin minimal promoter. Promoter induction was determined by luciferase assays 48 hr posttransfection (6).
RESULTS
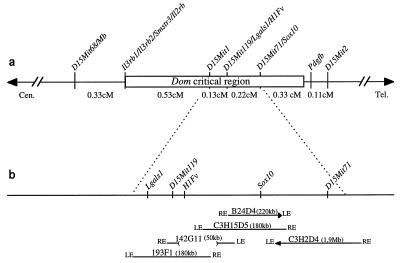
To be able to identify the Dom gene by positional cloning, we genetically mapped Dom to an interval of 1.6 cM between genetic markers D15Mit68 and D15Mit2 in a backcross with 252 Dom/+ progeny (15). This interval was further narrowed by increasing the progeny number, and by developing genetic markers that recombined on either side of the mutation, placing the Dom critical region between Il3rb2 and Pdgfb (Fig. 1a). We then constructed a YAC map of this region and analyzed several candidate genes contained in it (Fig. 1b). One of these genes was Sox10 (6). Sox10 cosegregated with D15Mit71 in the EUCIB backcross and was contained in two overlapping YACs of the contig.
Figure 1.
(a) Genetic map of the Dom region in the central-terminal part of chromosome 15. Genetic distances are given based on the EUCIB backcross. (b) Physical map of the the Dom region as determined from positive YACs. LE, left end; RE, right end. YAC chimerism is represented by an arrow.
The expression pattern of Sox10 (6) and the fact that it did not recombine with the Dom mutation prompted us to analyze this gene in greater detail. No gross alteration of the Sox10 gene was evident in Dom mice, and Sox10 transcripts could be detected. Direct sequencing of these transcripts revealed two differences compared with Sox10 transcripts from C57BL/6J mice (Fig. 2a). An A-to-T transversion at position 32 of the ORF caused a substitution of glutamate11 by valine, whereas the insertion of an additional G after position 579 resulted in an altered reading frame, which leaves the first 193 amino acids of Sox10 including the HMG-domain intact, but replaces the remaining 273 residues by a divergent carboxyl terminus of 99 unrelated amino acids.
Figure 2.
(a) Predicted amino acid sequences of wild-type (C57BL/6J) and mutant Sox10 (Dom). Divergent amino acids are set off by bold type; the DNA-binding HMG-domain is boxed. (b) Analysis of Sox10 expression by Northern blotting. Ten micrograms of total RNA from brain and intestine of newborn wild-type (+/+), heterozygous (Dom/+), and homozygous (Dom/Dom) mice were transferred to filters and sequentially hybridized with probes for Sox10 (Upper) and β-actin (Lower). Molecular sizes are indicated on the left.
Sox10 expression was analyzed in Dom/Dom pups obtained immediately after birth. In brain, which represents a major site of Sox10 expression at this developmental stage (6), transcript levels in homozygous Dom mutants were not reduced in comparison to those in heterozygous or wild-type littermates (Fig. 2b). In contrast, striking differences were observed in the intestine where Sox10 is expressed in the neural crest-derived cells (16) of the ENS (6). Here, Sox10 transcript levels were reduced in heterozygotes, and below detection in the newborn homozygous Dom mice, corresponding to a more severe manifestation of the defect in homozygous Dom mice in which the ENS fails to develop in the entire bowel below the rostral foregut (unpublished observation).
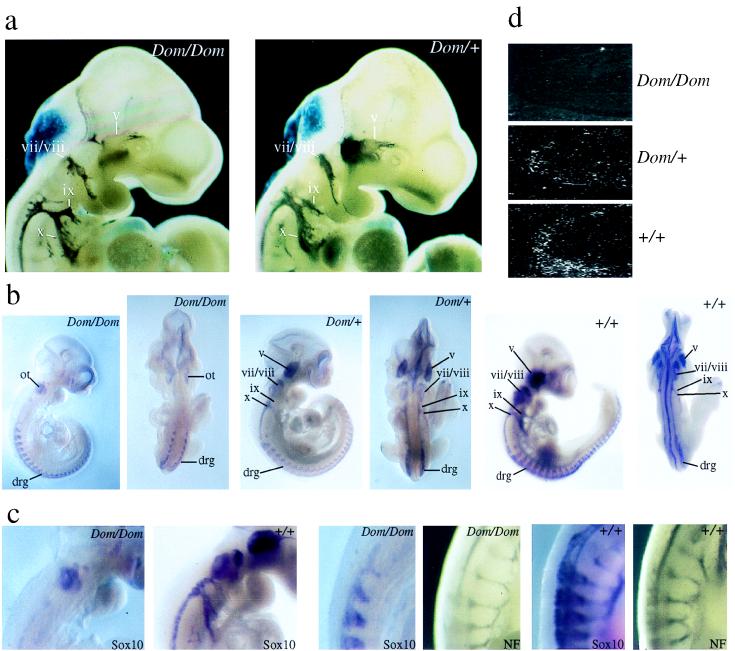
The combination of intestinal aganglionosis and pigmentation defects in the Dom mutant indicates that several neural-crest derived lineages are affected. Taking also the early embryonic lethality of many homozygous animals into account, this is best explained by a defect in early neural crest development. At E8.5, we failed to detect reproducible differences between neural crest cells of homozygous Dom mice and their wild-type littermates (data not shown). However, already at E10.5, striking alterations in the morphology of cranial nerves and ganglia were apparent in homozygous Dom mice after immunohistochemistry with specific antibodies against neurofilament protein (NF) (Fig. 3a). These alterations were reminiscent of the changes obtained by targeted deletion of neuregulin and erbB2 genes (17, 18), and were not detected in heterozygote Dom mice. In homozygous Dom mice, NF immunoreactivity was severely diminished in the dorsal portion of the trigeminal ganglion (v). The proximal portions of the glossopharyngeal (ix) and vagus (x) were considerably thinner than in heterozygote and wild-type mice, suggesting that the development of superior and jugular ganglia also was perturbed. Morphological alterations in the facial ganglion (vii) were restricted to the proximal portion of the nerve root. All affected structures predominantly contain neurons derived from the cranial neural crest, whereas those structures without detectable abnormalities are mostly placodal in origin (19, 20). No defect was detected in neurons of the trunk neural crest at E10.5, as NF immunoreactivity of dorsal root ganglia and motor nerves appeared unaltered.
Figure 3.
(a) Morphological comparison of cranial ganglia in homozygous mutant (Dom/Dom) and heterozygous (Dom/+) embryos at E10.5 by immunohistochemistry using anti-NF160 specific antibodies. Note that at this developmental stage, NF160 is expressed mainly in the peripheral nervous system. Cranial ganglia (trigeminal, v; facial/acoustic, vii/viii; glossopharyngeal, ix; and vagus, x) are marked. (b) Analysis of Sox10 expression in homozygous mutant (Dom/Dom), heterozygous (Dom/+), and wild-type (+/+) embryos at E10.5 by whole-mount in situ hybridization. Lateral and dorsal views are shown for each embryo. Localization of cranial ganglia (v, vii/viii, ix, x) and dorsal root ganglia (drg) are indicated. Hybridization in the otic vesicle (ot) was nonspecific. (c) Higher magnification of homozygous mutant (Dom/Dom) and wild-type (+/+) E10.5 mouse embryos to show Sox10 expression in cranial ganglia (left two panels) and to compare Sox10 expression (Sox10) with NF160 immunoreactivity (NF) in motor nerves (right four panels). (d) Sox10 expression in the intestine at E14.5 studied by in situ hybridization on sections of homozygous mutant (Dom/Dom), heterozygous (Dom/+), and wild-type (+/+) embryos.
Previous results had shown that Sox10 is itself expressed in cranial and trunk neural crest cells, and is later confined in the peripheral nervous system to glial cells (6). Whole-mount in situ hybridization showed that Sox10 expression was lost from all cranial ganglia and nerves of homozygous Dom mice at E10.5 (Fig. 3 b and c). The absence of Sox10-specific signals from these structures could result from either disregulation of Sox10 expression in cranial neural crest cells, or from the absence of Sox10 expressing cells. We currently favor the latter hypothesis because preliminary studies failed to detect any evidence of disregulation in tissue culture transfection systems (data not shown).
In contrast to cranial ganglia, Sox10 expression was detectable in the trunk neural crest-derived dorsal root ganglia (Fig. 3b). A substantial reduction in the intensity of the Sox10 hybridization signal was again observed along motor nerves (Fig. 3c). No qualitative differences were detected regarding the Sox10 expression pattern between heterozygous and wild-type mice at E10.5 (Fig. 3b). There was, however, an overall reduction of Sox10 expression in Dom/+ mice.
In situ hybridization of sections from E14.5 embryos allowed analysis of Sox10 expression in the gut at times when the ENS starts to develop (Fig. 3d). The majority of cells within the ENS are derived from the cranial (previously also referred to as vagal) neural crest (21) and transiently express Sox10 during embryogenesis (6). Contrary to wild-type mice, no Sox10-expressing cells were detected in the intestine of homozygous Dom mice, whereas few can be seen in the intestine of heterozygotes (Fig. 3d). This absence or reduction of Sox10 expression in the gut of homozygous and heterozygous Dom mice establishes a strong link between Sox10 and the defective colonization of the intestine by cranial neural crest that defines the Dom phenotype.
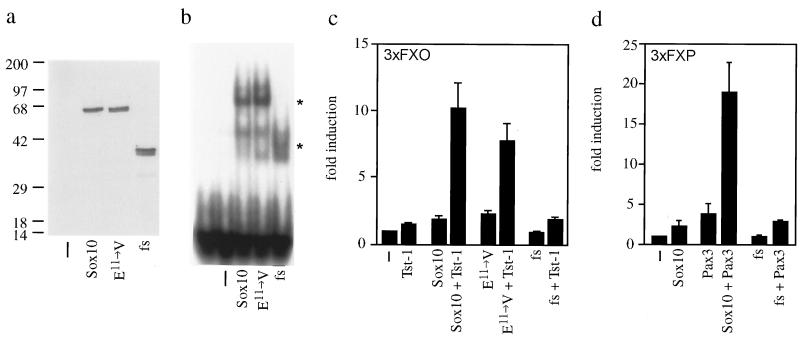
To analyze the functional consequences of the mutation present in the Sox10 gene of Dom mice on a molecular level, we separately introduced the glutamate11-to-valine substitution and the frameshift into the rat Sox10 cDNA. We chose the rat cDNA as it had been used in all previous functional studies on Sox10 (6). Furthermore, rat and mouse cDNA sequences are 96% identical over the complete ORF, and rat and mouse proteins even share 99% of their amino acids. Thus it is reasonable to assume that mouse and rat proteins behave identically. After transient transfection of COS cells, both mutant proteins were expressed at levels similar to wild-type Sox10 and were correctly localized to the cell nucleus as judged by their presence in nuclear extracts (Fig. 4a). Consistent with an unaltered HMG-domain, both mutant proteins bound to a consensus DNA recognition motif for Sox proteins with affinities comparable to the wild-type protein in electrophoretic mobility-shift assays (Fig. 4b).
Figure 4.
(a) Detection of Sox10 proteins (wild type, Sox10; the glutamate11-to-valine substitution mutant, E11→ V; and the Dom frameshift mutant, fs) in nuclear extracts of transiently transfected COS cells by Western blotting using a rabbit antiserum against Sox10. −, mock-transfected. (b) DNA binding of Sox10 proteins as determined by electrophoretic mobility-shift assay using a radioactively labeled consensus binding site for Sox proteins as probe and COS nuclear extracts from a as protein source. Sox10-specific complexes are marked by asterisks. (c) Promoter induction after transient transfection of the luciferase reporter plasmid 3xFXO luc into U138 glioblastoma cells in combination with empty CMV expression plasmid (−), pCMV/Tst-1 (Tst-1), and CMV expression plasmids for Sox10 proteins (Sox10, E11→ V, and fs). (d) Promoter induction after transient transfection of the luciferase reporter plasmid 3xFXP luc into U138 cells in combination with empty CMV expression plasmid (−), pCMV/Pax3 (Pax3), and CMV expression plasmids for Sox10 proteins. Results in c and d show mean (+SE) promoter induction (n = 4).
Previous studies had shown that Sox10 was not a strong transcriptional activator, but synergistically enhanced the activity of Tst-1/Oct6/SCIP and Pax3, two other transcription factors present in neural crest-derived cells at various developmental stages (6). The frameshift mutation after position 579 completely abolished this ability as evidenced in transient transfections of U138 glioblastoma cells (Fig. 4 c and d). In contrast, the glutamate11-to-valine substitution mutant synergistically enhanced transcriptional activity of Tst-1/Oct6/SCIP to the same extent as wild-type Sox10.
DISCUSSION
Our results define Sox10 as an important regulator of neural crest development. Its inactivation affects the early development of neural crest cells either in multipotent precursors before lineage restriction or thereafter in multiple neural crest lineages. As a consequence, inactivation of Sox10 leads to a loss not only of peripheral nervous system neurons and glia, but also to a loss of enteric ganglia, and judged by the pigmentation defect of Dom mice (3), to a loss of certain melanocyte populations. It is particularly intriguing that the cranial neural crest is more severely affected than the trunk neural crest. Thus, Sox10 might be involved in defining the regional identity of the cranial neural crest.
Furthermore, Sox10 is the first member of the Sox gene family shown to have an essential function in neural development, thus attributing an additional role to this group of transcriptional regulators, which so far have been mainly associated with other developmental processes such as male sex determination, hemopoiesis, and chondrogenesis and with corresponding defects such as XY sex reversal and campomelic dysplasia (22–26).
Similar to the ls and sl mutants, the Dom mutant also has been used in the past as a model system for Hirschsprung disease (HSCR). HSCR is a congenital human disease characterized by a lack of intestinal motility and a complete absence of neural crest-derived enteric ganglia in distal bowel (27). As both the endothelin-3 gene and the endothelin-B receptor gene, which are inactivated in the ls and sl mutants (4, 5), have been shown to be involved in HSCR (28), participation of Sox10 in HSCR is expected. It also deserves to be noted that the Dom phenotype combines intestinal aganglionosis with other neural crest defects that are highly reminiscent of the clinical features observed in Waardenburg syndrome (29). Thus, mutations of the human Sox10 gene might be present among patients with combined Hirschsprung and Waardenburg syndromes.
Acknowledgments
We thank R.M. Nitsch for advice on the manuscript, and I. Poras and P. Avner for their help at the Genethon Mouse YAC screening service. This study was supported by grants from the Deutsche Forschungsgemeinschaft and the Bundesministerium für Bildung, Wissenschaft, Forschung, und Technologie (to M.W.) and by grants from the Institut National de la Santé et de la Recherche Médicale and the French Ministry of Research (to M.G.). V.P. is a recipient of an Association Française Contre les Myopathies fellowship, and A.P. received an Italian Telethon fellowship.
ABBREVIATIONS
- E
embryonic day
- ENS
enteric nervous system
- HMG
high-mobility group
- YAC
yeast artificial chromosome
- NF
neurofilament protein
Note
: While this manuscript was under review, a similar report on the molecular basis of the Dom mouse was published (30).
Footnotes
References
- 1.Anderson D. Trends Genet. 1997;13:276–280. doi: 10.1016/s0168-9525(97)01187-6. [DOI] [PubMed] [Google Scholar]
- 2.Lane P W. J Hered. 1966;57:29–31. doi: 10.1093/oxfordjournals.jhered.a107457. [DOI] [PubMed] [Google Scholar]
- 3.Lane P W, Liu H M. J Hered. 1984;75:435–439. doi: 10.1093/oxfordjournals.jhered.a109980. [DOI] [PubMed] [Google Scholar]
- 4.Baynash A G, Hosoda K, Giaid A, Richardson J A, Emoto N, Hammer R E, Yanagisawa M. Cell. 1994;79:1277–1285. doi: 10.1016/0092-8674(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 5.Hosoda K, Hammer R E, Richardson J A, Baynash A G, Cheung J C, Giaid A, Yanagisawa M. Cell. 1994;79:1267–1276. doi: 10.1016/0092-8674(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 6.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pevny L H, Lovell-Badge R. Curr Opin Genet Dev. 1997;7:338–344. doi: 10.1016/s0959-437x(97)80147-5. [DOI] [PubMed] [Google Scholar]
- 8.Prior H M, Walter M A. Mol Med. 1996;2:405–412. [PMC free article] [PubMed] [Google Scholar]
- 9.Pingault V, Puliti A, Prehu M-O, Samadi A, Bondurand N, Goossens M. Genomics. 1997;39:86–89. doi: 10.1006/geno.1996.4476. [DOI] [PubMed] [Google Scholar]
- 10.Chartier F L, Keer J T, Sutcliffe M J, Henriques D A, Mileham P, Brown S D. Nat Genet. 1992;1:132–136. doi: 10.1038/ng0592-132. [DOI] [PubMed] [Google Scholar]
- 11.Larin Z, Monaco A P, Lehrach H. Proc Natl Acad Sci USA. 1991;88:4123–4127. doi: 10.1073/pnas.88.10.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossi J M, Burke D T, Leung J C, Koos D S, Chen H, Tilghman S M. Proc Natl Acad Sci USA. 1992;89:2456–2460. doi: 10.1073/pnas.89.6.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson P. Mamm Genome. 1995;6:429–432. doi: 10.1007/BF00355646. [DOI] [PubMed] [Google Scholar]
- 14.Mark M, Lufkin T, Vonesch J L, Ruberte E, Olivo J C, Dolle P, Gorry P, Lumsden A, Chambon P. Development (Cambridge, UK) 1993;119:319–338. doi: 10.1242/dev.119.2.319. [DOI] [PubMed] [Google Scholar]
- 15.Puliti A, Prehu M O, Simon-Chazottes D, Ferkdaji L, Peuchmayr M, Goossens M, Guenet J L. Mamm Genome. 1995;6:763–768. doi: 10.1007/BF00539000. [DOI] [PubMed] [Google Scholar]
- 16.Le Douarin N M. The Neural Crest. Cambridge, U.K.: Cambridge Univ. Press; 1982. [Google Scholar]
- 17.Lee K-F, Simon H, Chen H, Bates B, Hung M-C, Hauser C. Nature (London) 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 18.Meyer D, Birchmeier C. Nature (London) 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- 19.D’Amico-Martel A, Noden D M. Am J Anat. 1983;166:445–468. doi: 10.1002/aja.1001660406. [DOI] [PubMed] [Google Scholar]
- 20.Le Douarin N M, Smith J. Annu Rev Cell Biol. 1988;4:375–404. doi: 10.1146/annurev.cb.04.110188.002111. [DOI] [PubMed] [Google Scholar]
- 21.Gershon M D. Curr Opin Neurobiol. 1997;7:101–109. doi: 10.1016/s0959-4388(97)80127-4. [DOI] [PubMed] [Google Scholar]
- 22.Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Münsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. Nature (London) 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 23.Koopman P, Münsterberg A, Chapel B, Vivian N, Lovell-Badge R. Nature (London) 1990;348:450–452. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- 24.Foster J W, Dominguez-Steglich M A, Guioli S, Kwok C, Weller P A, Stevanovic M, Weissenbach J, Mansour S, Young I D, Goodfellow P N, et al. Nature (London) 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 25.Schilham M W, Oosterwegel M A, Moerer P, Ya J, Deboer P A J, van de Wetering M, Verbeek S, Lamers W H, Kruisbeek A M, Cumano A, Clevers H. Nature (London) 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 26.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli F D, Keutel J, Hustert E, et al. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 27.Badner J A, Sieber W K, Garver K L, Chakravarti A. Am J Hum Genet. 1990;46:568–580. [PMC free article] [PubMed] [Google Scholar]
- 28.Hofstra R M W, Osinga J, Buys C H C M. Eur J Hum Genet. 1997;5:180–185. [PubMed] [Google Scholar]
- 29.Read A P, Newton V E. J Med Genet. 1997;34:656–665. doi: 10.1136/jmg.34.8.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Southard-Smith E M, Kos L, Pavan W J. Nat Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]