Abstract
Although it is well-established that p53 functions as a tumor suppressor gene, certain mutations exhibit gain-of-function activities that increase oncogenic transformation. We have found a common class of p53 missense mutation that exhibits a dominant, gain-of-function activity that generates genomic instability. Fibroblasts from Li–Fraumeni syndrome heterozygotes with such mutations generate polyploid cells when exposed to spindle depolymerizing agents. Expression of such mutant alleles in normal fibroblasts yields the same phenotype. This class of dominant, gain-of-function p53 mutation (p53RSC, relaxed spindle checkpoint allele) does not require the transcriptional activation function of p53 for this behavior. Thus p53 mutations can contribute to progression of a cancer cell not only by absence of p53 tumor suppressor activity but also by the presence of an activity that promotes genetic instability.
The p53 gene is one of the most frequently altered genes in a wide variety of tumor cells (reviewed in ref. 1), indicating that it is important in growth control and tumorigenesis. Understanding how mutations in p53 contribute to neoplastic transformation is under intensive investigation (2, 3). The majority of p53 mutations apparently result in loss of function. One way in which loss of p53 activity can occur is through truncation or deletion of both wild-type alleles in diploid cells. Mice that are homozygous for deletion of both p53 alleles exhibit increased tumor incidence and provide examples of such loss-of-function mutations (4, 5). Likewise, the loss of wild-type p53 activity in tissue culture cells removes important controls on cell cycle regulation, apoptosis, and maintenance of genomic integrity (6) and contributes to tumor development (7). Although deletion of the gene and concomitant loss of wild-type p53 function clearly contribute to tumorigenesis, missense mutations in p53 also may lead to a loss of function by generating a dominant negative form that inhibits the activity of wild-type p53 (8). In this case, expression of a dominant negative mutant p53 would result in a phenotype that is indistinguishable from that seen in p53 null cells. Such mutations have been found and contribute to the tumorigenic phenotype (9–11).
In principle, missense mutations also can contribute to tumorigenesis by creating a novel gain-of-function form. A gain-of-function mutation of this sort can be distinguished from a dominant negative mutation because it causes a novel phenotype that is not seen in the p53 null cell. An indication that a p53 mutation can promote tumorigenesis above the level seen in p53 null cells was first described by Wolf et al. (12), in which the expression of a mutant p53 in a p53 null cell enhanced malignant transformation. Additional reports have come from several laboratories demonstrating gain-of-function activities that affect tumor progression (13–18).
Several reports support a role for mutant p53 in the generation of aneuploidy in human cells. An accumulation of aneuploid cells has been found in vitro in fibroblasts from Li–Fraumeni syndrome (LFS) patients, who carry a congenital mutation in one p53 allele (19). Moreover, the expression of mutant p53 proteins in human colon carcinoma cells results in a tendency to increase ploidy level during growth in culture (20) or in response to radiation or adriamycin treatment (21). To understand how the presence of mutant p53 proteins might affect cell cycle control at mitosis in preneoplastic human cells, we investigated the cellular response to spindle inhibitors of normal human fibroblasts (NHFs) and fibroblasts from apparently normal skin biopsies of members of LFS families. The LFS cell populations included in this study were selected because they represent a variety of p53 mutations that fall into three categories: (i) truncations that completely inactivate p53, (ii) mutations in the site-specific DNA binding domains of p53 (class I mutants) that disrupt the binding of the protein to a DNA consensus sequence (22), and (iii) p53 conformational mutations (class II mutants) that affect residues that contribute to the maintenance of the tertiary structure or conformation of the protein (22, 23). LFS heterozygotes carrying these three types of p53 mutations were examined for spindle checkpoint control. Some of these mutations (most notably in class II) demonstrated altered spindle checkpoint control and underwent repeated rounds of DNA synthesis without chromosomal segregation, generating polyploid cells.
MATERIALS AND METHODS
Characterization of LFS Heterozygotes.
Cells and their p53 genotype are indicated in Table 1. Origin and propagation of cells are described in ref. 24. Heterozygosity was verified by sequencing, antibody staining and a yeast-based gene expression assay (Oncor). Production of the truncated p53 protein in JML cells was visualized by Western analysis using Ab DO1. Mutant p53 protein expression was detected in SF and 174 cells by immunostaining with Pab240. All cell populations were routinely monitored for mycoplasma infection and found to be negative.
Table 1.
Characterization of LFS heterozygotes
| Cell population | Passage | Cell doubling time, h | p53 genotype | Ploidy in colcemid | Class of mutation |
|---|---|---|---|---|---|
| NHF | p9 | 20 | +/+ | 4n | — |
| SF | p9 | 44 | +/248W | 4n | Class I |
| O41 | p17 | 60 | +/184T | 4n | Truncation |
| JML | I* | 30 | 184T/184T | 4n | Truncation |
| 174 | p12 | 21 | +/175H | Polyploid | Class II |
| 4404 | p11 | 39 | +/175H | Polyploid | Class II |
| 9136 | p18 | 77 | +/245D | Polyploid | Class II |
| 143A | p11 | 26 | +/+/143A | Polyploid | Class II |
| 175H | p8 | 25 | +/+/175H | Polyploid | Class II |
| 281G | p11 | 24 | +/+/281G | Polyploid | Class II |
Immortal.
Transfections and Infections.
Infections of NHFs with retroviruses producing human papilloma virus (HPV) type 16 E6 protein were as previously described (25). Retroviral vectors expressing p53–143A mutant protein were created by subcloning a BamHI cDNA fragment encoding p53–143A in the retroviral vector pBabe (puro) (26). Plasmids containing p53–281G or p53–281G,22,23, and the neo resistance gene sequences were transfected by the calcium phosphate method into NHFs followed by drug selection.
Flow Cytometry.
Fibroblasts were processed as in ref. 25. Briefly, all cells are pulsed with BrdUrd for 4 hr just before harvesting to label cells that are synthesizing DNA. After isolation of the nuclei, the samples are counterstained with propidium iodide (PI), which allows for the determination of total DNA content in each nucleus. Reaction with an antibody that detects BrdUrd and separation by flow cytometry allows the separation of nuclei into populations containing G1, S, G2/M, and >G2 contents of DNA. The data are either exhibited as a three-dimensional plot as in Fig. 1 or as a two-dimensional plot of cell number versus PI concentration as in Figs. 2–4.
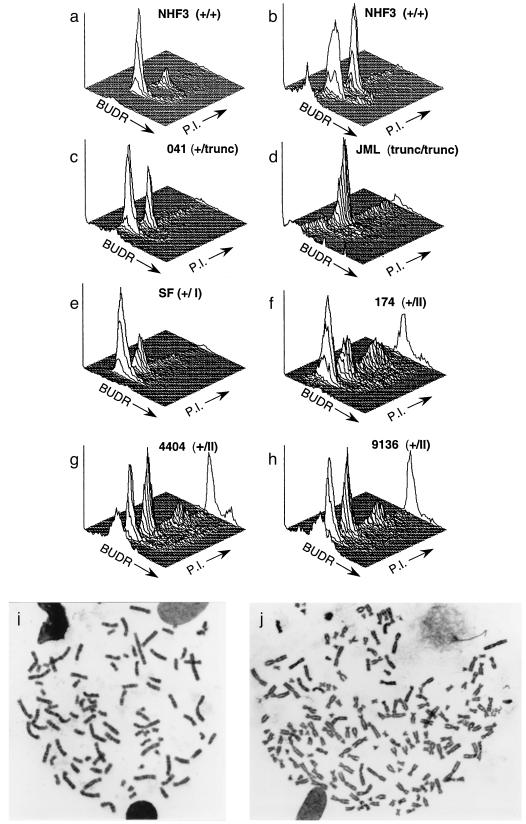
Figure 1.
Analysis of the status of the spindle cell cycle checkpoint in human fibroblasts. Cell cycle was analyzed by flow cytometry to determine the distribution of DNA content in NHF3 incubated without (a) or with (b) 500 ng/ml of colcemid for two population doublings. With increasing time in colcemid, an increasing fraction of normal cells accumulate in the G2/M peak. LFS fibroblasts (c–h) are described in Table 1 and ref. 24. LFS cells were processed for cell-cycle distribution of DNA content as for NHF, except for JML cells, which were incubated for 1 population doubling time. The three-dimensional plots represent cell number (y axis) versus BrdUrd (BUDR) incorporation (z) and PI (x). Plots show 104 cells. (i and j) Metaphase spreads from NHF cells expressing a p53RSC mutation (p53–143A) after two population doubling times in colcemid. Metaphase spreads with 92 (i) and 182 (j) chromosomes are presented.
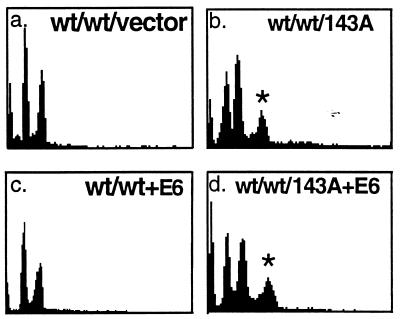
Figure 2.
The expression of a p53RSC (p53–143A) in NHFs (NHF29) alters the spindle cell cycle checkpoint. Flow cytometric analysis was performed to obtain the cell cycle distribution of DNA content in colcemid-treated NHFs infected with retroviruses that produce p53–143A mutant protein (b), HPV16 E6 protein (c), both p53–143A and HPV16 E6 proteins (d), or neither (a). Polyclonal populations at passage 1 were incubated and processed for cell cycle distribution of DNA content as in Fig. 1. Population doubling times for NHF29, NHF29-E6, NHF29-p53–143A, and NHF29-p53–143A+E6 were 30, 24, 30, and 36 h, respectively. wt refers to the presence of the wild-type allele, and ∗ indicates the polyploid population of cells. The x axis represents PI intensity and the y axis represents cell number.
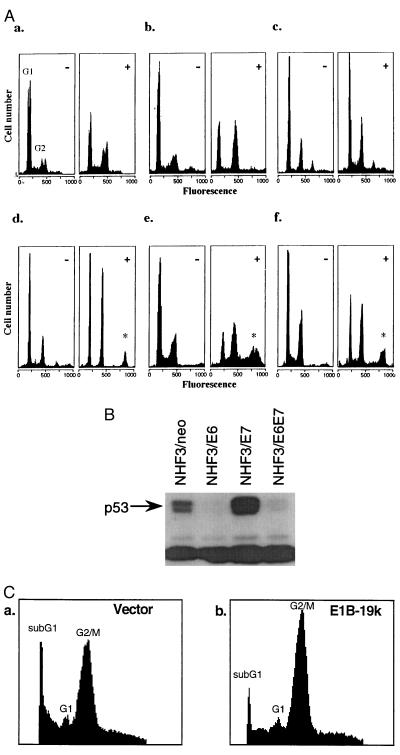
Figure 4.
(A) Loss of wild-type p53 activity in human cells does not generate polyploid cells. NHFs expressing HPV16 E6 protein [NHF1-E6 (a), NHF3-E6 (b), and NHF29-E6 (c)] were incubated without (−) or with (+) colcemid and analyzed as in Fig. 2. There is no change in the fraction of polyploid cells. Expression of the p53RSC 143A allelle in colcemid-treated cells produces polyploid populations (denoted by ∗) [NHF3–143A (d), NHF29–143A (e), and FF6–143A (f)]. (B) Western analysis of p53 protein in NHF3 fibroblasts after expression of an empty vector containing the neomycin resistance gene, or the HPV16 oncoproteins E6, E7, or E6/E7 together. (C) Production of E1B 19k affects cell death but not ploidy in human p53 null cells. JML cells were stably transfected by the calcium phosphate method with plasmids containing the neo resistance gene alone (a) or with E1B 19k (b). Drug-resistant colonies were pooled and subject to colcemid treatment for two population doubling times (48 h), and analyzed for DNA content by PI staining.
Metaphase Spreads.
Spreads were processed as in ref. 25.
DNA Sequencing.
Sequencing was performed as previously described (24).
RESULTS
Spindle Checkpoint Activity in NHFs and LFS Heterozygotes.
NHFs incubated with colcemid (Fig. 1b) arrest with 4n DNA content. The spindle-dependent cell-cycle arrest was accompanied by a decrease in G1 cells (59% to 30%) and an increase in G2/M cells (21% to 39%) when compared with log-phase NHF cells in the absence of colcemid (Fig. 1a). Colcemid did not affect the progression of NHF3 cells throughout S phase. Approximately 10% of NHF3 cells were in S phase when incubated with or without colcemid.
We next examined LFS fibroblasts for the activation of spindle checkpoint control. LFS fibroblasts heterozygous for either class I mutations (Fig. 1e, SF) or truncation of wild-type p53 (Fig. 1c, 041) exhibited a normal spindle checkpoint response in which cells accumulated with 4n content of DNA (Table 1) when incubated in colcemid (30% and 33%, respectively). These populations were verified as heterozygotes by using several assays for expression of wild-type p53 activity (see Materials and Methods). JML cells, an immortal derivative of 041 cells, which are homozygous for the p53 truncation, also accumulate with 4n DNA content when incubated with colcemid (Fig. 1d).
Strikingly, cells heterozygous for a class II mutation (Fig. 1f, 174) exhibit a disrupted spindle checkpoint control. Instead of arresting with 4n DNA content, these cells re-entered S phase at G2 and underwent a new round of DNA replication that resulted in the appearance of cells with a polyploid DNA content. An average of 20% of the population of 174 cells reached an 8n DNA content when incubated in colcemid for the time normally required for two population doublings. Additional time in colcemid resulted in the generation of cells with even higher ploidy content of DNA such as 16n, 32n, etc. (data not shown). Two additional LFS cell populations from independent kindreds (4404 and 9136) demonstrated the same phenotype (Fig. 1 g and h, Table 1). The presence of cells with a polyploid DNA content was confirmed by karyotypic analysis (Fig. 1 i and j). Incubation of cells carrying the 175H mutation with nocodazole, another spindle assembly inhibitor, but not with the DNA synthesis inhibitors hydroxyurea or thymidine, demonstrated S-phase re-entry similar to that seen with colcemid (not shown). Cells with even greater amounts of DNA can be detected in Fig. 1 f–h as shown by the points at the extreme of the PI axis. These results indicated that LFS fibroblasts carrying specific p53 mutations (p53–175H or p53–245D), but not p53–248W mutations or truncations of p53, alter the spindle cell cycle checkpoint and generate cells with a polyploid DNA content. We refer to these alleles of p53 as p53RSC to reflect the phenotype of a relaxed spindle checkpoint.
S-Phase Re-Entry Is a Dominant Gain-of-Function Phenotype.
We have demonstrated that LFS cells, heterozygous for p53RSC mutations, exhibit a defect in spindle checkpoint control. If the p53 mutation in these cells is responsible for this behavior, we predict that introduction of such mutations into wild-type cells will exert a dominant phenotype when expressed in NHF cells. In other words, S-phase re-entry will occur in the presence of the wild-type gene when the cells are incubated in colcemid. Introduction of empty retroviral vectors (Fig. 2a) or vectors expressing excess wild-type p53 (data not shown) did not abrogate the spindle checkpoint control and resulted in cells that were arrested with 4n DNA content on incubation with colcemid. In contrast, NHFs infected with retroviral vectors encoding a variety of p53RSC mutants (143A, 281G, or 175H alleles) displayed DNA contents that were polyploid when these cells were incubated with colcemid (Figs. 2b and 3B and data not shown).
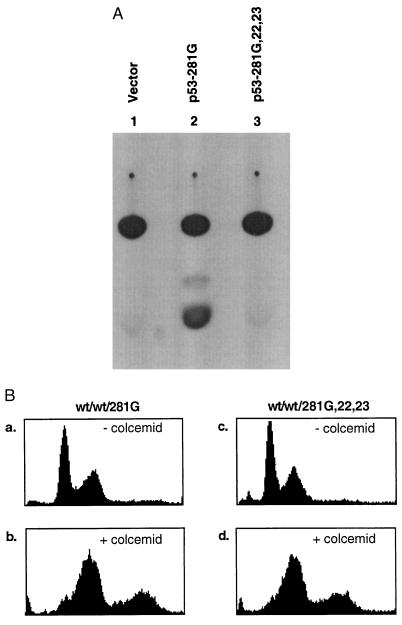
Figure 3.
Characterization of reporter activity and spindle cell cycle checkpoint induced by a transcription-deficient p53 mutant protein. (A) NHF3 cells were transiently cotransfected with 5 μg of MDR1 CAT reporter plasmid and 10 μg of either p53–281G or p53–281G,22,23 using Lipofectamine. After 48 h, cells were lysed and CAT activity was assayed using 14C-labeled chloramphenicol (31). CAT activity was observed only in cells cotransfected with the reporter alone nor cells cotransfected with p53-281G,22,23, and MDR1 CAT (lanes 1 and 3). (B) Flow cytometric analysis of NHF stably transfected with either p53–281G or p53–281G,22,23 in the presence (b and d) or absence (a and c) of colcemid. Colonies expressing mutant p53 were pooled and processed for cell cycle distribution of DNA content after colcemid treatment for 48 h as described in Fig. 1. Flow cytometry documented a decrease in apoptotic cells but no increase in polyploid cells.
To distinguish a dominant negative phenotype from a gain-of-function phenotype, we also expressed the p53–143A mutation in NHF3 cells that have p53 inactivated through coexpression of the HPV16 E6 oncoprotein. This oncoprotein promotes the degradation of wild-type but not mutant p53 (25, 27) and the decrease in wild-type p53 protein is shown in Fig. 4B by Western analysis. If the p53RSC mutations are dominant negative, the phenotype in p53 null cells should be identical to those carrying the p53RSC mutations. However, if the p53RSC mutations exhibit a gain-of-function phenotype, the phenotype will differ from the phenotype seen in the p53 null cells. The generation of polyploidy was observed in cells carrying the p53RSC mutation in the absence of wild-type p53 (Fig. 2d) but not in the population of cells that expressed E6 alone (Fig. 2c). These results indicate that the accumulation of a polyploid DNA content in human fibroblasts heterozygous for the p53RSC mutation is a gain-of-function phenotype that results from the expression of p53RSC mutant proteins and is not simply caused by inactivation of p53.
S-Phase Re-Entry Phenotype Is Independent of Transcriptional Activation by p53.
It has been postulated that the integrity of the p53 transactivation domain may be required for the gain-of-function properties of mutant p53 proteins (28). We tested whether the generation of polyploidy was one of them. Previously, the single mutant p53–281G, but not the triple mutant p53–281G,22,23, has been shown to transactivate the MDR1 promoter in both p53-deficient mouse fibroblasts and in human osteosarcoma (SAOS-2) cells (29). To confirm that a similar transactivation of the MDR1 promoter could occur in NHFs, NHF cells, transiently transfected with either p53–281G or p53–281G,22,23, were cotransfected with an MDR1 chloramphenicol acetyltransferase (CAT) reporter plasmid and tested for their ability to activate transcription of the MDR1 promoter. As reported previously (29), CAT activity was observed only in cells expressing p53–281G, but not in cells expressing p53–281G,22,23 (Fig. 3A). We then tested whether mutation of these two residues affected the ability of a p53RSC mutant, p53–281G, to promote the acquisition of polyploidy in colcemid-treated NHF. When these cell populations were exposed to colcemid, they showed similar degrees of polyploidy (compare Fig. 3B b and d, 24% for both), indicating that loss of the transactivation activity by this p53 mutant does not affect the ability of these proteins to alter the spindle cell cycle checkpoint. Transcription-independent p53 functions have been previously described (30).
Difference in Checkpoint Control Between Murine and Human Cells.
Previous studies have reported that murine cells devoid of wild-type p53 activity generate polyploid cells when incubated with spindle inhibitors (32). We repeated the murine experiments by using the p53 knockout mouse embryo fibroblasts described previously (24) and obtained results identical to those reported by Reid and coworkers (32) (data not shown); i.e., cells that were null for p53 did not arrest with a 4n DNA content but generated polyploid cells. As noted above, we found that human cells lacking p53 activity and incubated in colcemid arrested with 4n DNA content, a result that differs from the murine studies (Fig. 1d, JML cells; Fig. 2c). To confirm the distinction between mouse and human cells, we examined human cells that lacked wild-type p53 through three different mechanisms. First, using retroviral infection, we generated several NHF cell populations that express the HPV16 E6 protein, which promotes the degradation of wild-type p53 (32). Negligible quantities of p53 were observed by Western analysis in NHF3 cells expressing HPV16 E6, and a representative analysis is presented in Fig. 4B. We know that wild-type p53 activity is abolished in these cells because our previous experiments demonstrated that exposure to negative growth signals fails to activate G1 and G2 checkpoints (25). These cells, when incubated in colcemid, arrested with 4n DNA content. Four independent primary fibroblast populations were infected with HPV16 E6, and none demonstrated accumulation of polyploid cells over time in the presence of a spindle inhibitor (Figs. 2c and 4A, NHF1-E6, NHF3-E6, and NHF29-E6). These primary cell populations have the potential to produce polyploid cells as demonstrated by the expression of 143A p53RSC in each, and their generation of polyploid cells after exposure to colcemid Fig. 4A d–f (NHF3–143A, NHF29–143A, and FF6–143A). Although this experiment examines primary human cells that have lost wild-type p53 function, the conditions are not identical to those of the primary murine cells with a knockout of both p53 alleles. The HPV16 E6 oncoprotein has other functions besides binding p53 and theoretically could affect the generation of polyploid cells. These conclusions differ from those recently published (33), but differences in cells, mitotic inhibitors, method of analysis, and cell age may be responsible.
As a second approach, we examined human tumor cells (JML) that are homozygous for a p53 truncation and hence lack wild-type p53 protein. Incubation in colcemid resulted in the accumulation of cells with 4n DNA content (Fig. 1d). However, in these cells treatment with colcemid for periods longer than one population doubling time resulted in cell death. Visualization of condensed chromatin and fragmented nuclear morphology by H33258 staining and nucleosomal length DNA content by gel electrophoresis strongly suggested that these cells died by an apoptotic-like event (data not shown). Because the human p53 null cells suffered cell death after colcemid treatment, it is possible that acquisition of polyploidy was not detected because these cells re-enter S phase to be subsequently destroyed by a spindle checkpoint-dependent mechanism of cell death. We therefore determined whether inhibition of cell death by the expression of an apoptosis inhibitor, E1B 19K (34), would unmask polyploidy in human cells lacking p53 expression by decreasing cell death. Fig. 4C indicates that although the stable expression of E1B 19K in JML cells inhibited cell death after colcemid treatment (note the decrease in subG1 peak size from 22% to 10%) and allowed the visualization of cells that were arrested with 4n content of DNA (G2 increased from 53% to 63%), the fraction of polyploid cells remained the same (Fig. 4C a and b, 13% and 14%, respectively). These data indicate that these cells, which carry a homozygous p53 truncation mutation, cannot generate polyploid genomes even when cell death is inhibited.
Finally, in a third approach, we removed wild-type p53 activity in several NHF populations by expressing an antisense p53 sequence using retroviral vectors. No detectable p53 protein was observed in the antisense p53 cell populations as indicated by Western analysis (not shown). When these cell populations were analyzed for spindle cell cycle checkpoint control, we observed results that were similar to those seen with the JML cells described above: an increase in apoptosis. However, no increase in polyploidy was observed when the cells were incubated in colcemid (data not shown). Although it has been shown that the expression of high levels of an N-terminal portion of p53 by transient transfection can induce apoptosis (35), antisense experiments indicate that, in the case of the spindle cell cycle checkpoint in human cells, cell death results from a p53-independent mechanism. A colcemid-induced p53-independent apoptotic mechanism recently has been demonstrated in mouse cells (36). The differences between the antisense p53 and the E6 cell populations may be the result of the ability of the E6 protein to protect cells from the induction of cell death (37). In summary, removal of wild-type p53 expression by three different means in human cells all resulted in cells that retained an intact spindle checkpoint response. In contrast, removal of p53 in knockout mice or by SV40 T antigen expression in murine cells permits the generation of polyploid cells.
DISCUSSION
We have shown that both LFS heterozygotes and NHF cells that express p53RSC mutations exhibit a dominant, gain-of-function phenotype when the cells are incubated with a mitotic inhibitor. These data provide evidence for the action of mutant p53 as an oncogene that alters the maintenance of genomic integrity in human cells. The appearance of S-phase re-entry in human cells carrying p53RSC mutations may have drastic consequences in terms of genomic integrity, because polyploid fibroblasts have been shown to be genomically unstable, losing chromosomes randomly and generating aneuploidy (19, 38). This altered cell cycle checkpoint status may constitute an important step leading to the heterogeneity observed in tumors with p53 mutations (39) and tumor progression.
Previous work has highlighted the critical role of the p53 protein in cell cycle checkpoint control and its role in maintaining genomic integrity (24, 40). The onset of each phase of the cell cycle depends on the completion of events in the previous cell-cycle phase and the coordination of these events is maintained by cell-cycle checkpoints (41, 42). The present report describes a class of gain-of-function mutations in p53 that prevents the proper regulation of the spindle checkpoint and results in the generation of genomic instability, i.e., polyploidy. The generation of polyploid and aneuploid cells has been studied in yeast, and some components of the signal transduction pathways that oversee the successful propagation and segregation of chromosomes that prevent these chromosomal changes have been identified. Mutation in these genes result in chromosomal rearrangements, duplications, aberrant S-phase re-entry, and polyploidy (43–45).
Our studies have identified p53RSC alleles as containing missense mutations. Our limited analysis correlates the p53RSC phenotype with p53 mutations that have altered conformational structure (class II mutatation). However, a more comprehensive analysis of other missense mutations is needed before ruling out the possibility that certain class I mutations do not exhibit this dominant, gain-of-function phenotype. Most of the mutations of p53 observed in human tumors are missense mutants (81.4%) (7), and fully half of these missense mutations are class II mutants, suggesting that this alteration in cell-cycle checkpoint control may play a significant role in human tumorigenesis. The other types of mutations, such as mutants in the DNA-binding domain of p53, may affect additional phenotypes in more advanced stages of tumor progression once the wild-type allele is inactivated. In fact, most tumors with an altered p53 status contain a deleted p53 allele and a missense mutant allele (46–48).
The mechanism by which the p53RSC alleles promote the formation of polyploid cells is unknown. The phenotype could be produced by several different types of cell cycle malfunctions (49). Repeated rounds of S phase without entry into G2/M could produce the polyploid cells observed. We do not favor this mechanism, however, because polyploid cells with condensed chromosomes were observed (Fig. 1 i and j), indicating that chromosomal condensation, which occurs during M phase, was functional. Another possibility is that the expression of the p53RSC protein could allow for the inactivation of the spindle checkpoint itself or alter the timing of the adaptation response (50), which is known to override the checkpoint after a period of time. Mutations in the yeast adaptation response recently have been isolated (51). Our present studies would not distinguish between these two possibilities. Kinetic determinations of cell-cycle progression are underway to characterize the presence or duration of the spindle checkpoint response in cells expressing p53RSC in the presence of colcemid.
Although many aspects of metabolism and biology are conserved between species, some affecting neoplasia differ. It is not unexpected to find fundamental differences between murine and human cells. The frequencies of immortalization and neoplastic transformation differ dramatically between these two species (52). Spontaneous immortalization of murine cells is at least a million times more frequent in mouse than in human cells. Likewise, neoplastic transformation of murine cells after chemical exposure is also vastly different than in human cells. Although the chemical transformation of rodent cells is a routine experimental procedure, occurring with high frequency, the chemical transformation of human cells has yet to be accomplished in a reproducible manner. Finally, gross inspection of chromosomal spreads also document that genomic instability is much more prevalent in murine and other rodent species than in human cells (53). The basis for these observations is unknown. However, because alterations in the regulatory pathways that govern cell-cycle progression may contribute to each of the above processes, a difference between murine and human cells is important to investigate. Our results stress the importance of using human cells to study the role that certain alterations in cell-cycle checkpoint control may play in human tumor initiation/progression.
Acknowledgments
We thank J. Campisi, M. Hixon, H. Land, A. Levine, B. Vogelstein, and E. White for reagents. We thank S. Friend and M. Kastan for LFS fibroblasts. Insightful critique was provided by Drs. Dean Felsher, Ira Herskowitz, Charles Holst, Maxwell Heiman, and members of the Tlsty laboratory. This work was supported by National Institutes of Health Grants CA58207, CA51912, CA42765, and CA58207 to T.D.T.
ABBREVIATIONS
- LFS
Li–Fraumeni syndrome
- NHF
normal human fibroblast
- HPV
human papilloma virus
- PI
propidium iodide
- CAT
chloramphenicol acetyltransferase
References
- 1.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 2.Lane D P, Benchimol S. Genes Dev. 1990;4:1–8. doi: 10.1101/gad.4.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Hann B C, Lane D P. Nat Genet. 1995;9:221–222. doi: 10.1038/ng0395-221. [DOI] [PubMed] [Google Scholar]
- 4.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Nature (London) 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 5.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 6.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 7.Levine A J. Annu Rev Biochem. 1993;62:623–651. doi: 10.1146/annurev.bi.62.070193.003203. [DOI] [PubMed] [Google Scholar]
- 8.Herskowitz I. Nature (London) 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 9.Finlay C A, Hinds P W, Levine A J. Cell. 1989;57:1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 10.Srivastava S, Wang S, Tong Y A, Pirollo K, Chang E H. Oncogene. 1993;8:2449–2456. [PubMed] [Google Scholar]
- 11.Harvey M, Vogel H, Morris D, Bradley A, Bernstein A, Donehower L A. Nat Genet. 1995;9:305–311. doi: 10.1038/ng0395-305. [DOI] [PubMed] [Google Scholar]
- 12.Wolf D, Harris N, Rotter V. Cell. 1984;38:119–126. doi: 10.1016/0092-8674(84)90532-4. [DOI] [PubMed] [Google Scholar]
- 13.Eliyahu D, Michalovitz D, Oren M. Nature (London) 1985;316:158–160. doi: 10.1038/316158a0. [DOI] [PubMed] [Google Scholar]
- 14.Pohl J, Goldfinger N, Radler-Pohl A, Rotter V, Schirrmacher V. Mol Cell Biol. 1988;8:2078–2081. doi: 10.1128/mcb.8.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaulsky G, Goldfinger N, Rotter V. Cancer Res. 1991;51:5232–5237. [PubMed] [Google Scholar]
- 16.Dittmer D, Pati S, Zambetti G, Chu S, Teresky A K, Moore M, Finlay C, Levine A J. Nat Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 17.Hsiao M, Low J, Dorn E, Ku D, Pattengale P, Yeargin J, Haas M. Am J Pathol. 1994;145:702–714. [PMC free article] [PubMed] [Google Scholar]
- 18.Iwamoto K S, Mizuno T, Ito T, Tsuyama N, Kyoizumi S, Seyama T. Cancer Res. 1996;56:3862–3865. [PubMed] [Google Scholar]
- 19.Bischoff F Z, Yim S O, Pathak S, Grant G, Siciliano M J, Giovanella B C, Strong L C, Tainsky M A. Cancer Res. 1990;50:7979–7984. [PubMed] [Google Scholar]
- 20.Agapova L S, Ilyinskaya G V, Turovets N A, Ivanov A V, Chumakov P M, Kopnin B P. Mutat Res. 1996;354:129–138. doi: 10.1016/0027-5107(96)00062-0. [DOI] [PubMed] [Google Scholar]
- 21.Waldman T, Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1996;381:713–716. doi: 10.1038/381713a0. [DOI] [PubMed] [Google Scholar]
- 22.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 23.Prives C. Cell. 1994;78:543–546. doi: 10.1016/0092-8674(94)90519-3. [DOI] [PubMed] [Google Scholar]
- 24.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 25.White A E, Livanos E M, Tlsty T D. Genes Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 26.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munger K, Scheffner M, Huibregtse J M, Howley P M. Cancer Surv. 1992;12:197–217. [PubMed] [Google Scholar]
- 28.Levine A J, Chang A, Dittmer D, Notterman D A, Silver A, Thorn K, Welsh D, Wu M. J Lab Clin Med. 1994;123:817–823. [PubMed] [Google Scholar]
- 29.Lin J, Teresky A K, Levine A J. Oncogene. 1995;10:2387–2390. [PubMed] [Google Scholar]
- 30.Caelles C, Helmberg A, Karin M. Nature (London) 1994;370:220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 31.Doyle K, editor. Protocols and Applications Guide. 3rd Ed. Madison, WI: Promega; 1996. pp. 223–225. [Google Scholar]
- 32.Cross S M, Sanchez C A, Morgan C A, Schimke M K, Ramel S, Idzerda R L, Raskind W H, Reid B J. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 33.Di Leonardo A, Khan S H, Linke S P, Greco V, Seidita G, Wahl G M. Cancer Res. 1997;57:1013–1019. [PubMed] [Google Scholar]
- 34.White E, Sabbatini P, Debbas M, Wold W S, Kusher D I, Gooding L R. Mol Cell Biol. 1992;12:2570–2580. doi: 10.1128/mcb.12.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haupt Y, Rowan S, Shaulian E, Vousden K H, Oren M. Genes Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 36.Minn A J, Boise L H, Thompson C B. Genes Dev. 1996;10:2621–2631. doi: 10.1101/gad.10.20.2621. [DOI] [PubMed] [Google Scholar]
- 37.Pan H, Griep A E. Genes Dev. 1995;9:2157–2169. doi: 10.1101/gad.9.17.2157. [DOI] [PubMed] [Google Scholar]
- 38.Shackney S E, Smith C A, Miller B W, Burholt D R, Murtha K, Giles H R, Ketterer D M, Pollice A A. Cancer Res. 1989;49:3344–3354. [PubMed] [Google Scholar]
- 39.Carder P, Wyllie A H, Purdie C A, Morris R G, White S, Piris J, Bird C C. Oncogene. 1993;8:1397–1401. [PubMed] [Google Scholar]
- 40.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 41.Hartwell L H, Weinert T A. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 42.Elledge S J. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 43.Li R, Murray A W. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 44.Hoyt M A, Totis L, Roberts B T. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 45.Rudner A D, Murray A W. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- 46.Masuda H, Miller C, Koeffler H P, Battifora H, Cline M J. Proc Natl Acad Sci USA. 1987;84:7716–7719. doi: 10.1073/pnas.84.21.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nigro J M, Baker S J, Preisinger A C, Jessup J M, Hostetter R, Cleary K, Bigner S H, Davidson N, Baylin S, Devilee P. Nature (London) 1989;342:705–708. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- 48.Caron de Fromentel C, Soussi T. Genes Chromosomes Cancer. 1992;4:1–15. doi: 10.1002/gcc.2870040102. [DOI] [PubMed] [Google Scholar]
- 49.Kung A L, Sherwood S W, Schimke R T. Proc Natl Acad Sci USA. 1990;87:9553–9557. doi: 10.1073/pnas.87.24.9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinert T A, Hartwell L H. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toczyski D P, Hartwell L H. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- 52.Barrett J C, Tsutsui T, Tlsty T D, Oshimura M. Genetic Mechanisms in Carcinogenesis and Tumor Progression. New York: Wiley–Liss; 1990. pp. 97–114. [Google Scholar]
- 53.Yerganian G. In: Molecular Cell Genetics. Gottesman M M, editor. New York: Wiley; 1985. pp. 3–36. [Google Scholar]